Nitin Nitin
Enhancing AI microscopy for foodborne bacterial classification via adversarial domain adaptation across optical and biological variability
Nov 29, 2024Abstract:Rapid detection of foodborne bacteria is critical for food safety and quality, yet traditional culture-based methods require extended incubation and specialized sample preparation. This study addresses these challenges by i) enhancing the generalizability of AI-enabled microscopy for bacterial classification using adversarial domain adaptation and ii) comparing the performance of single-target and multi-domain adaptation. Three Gram-positive (Bacillus coagulans, Bacillus subtilis, Listeria innocua) and three Gram-negative (E. coli, Salmonella Enteritidis, Salmonella Typhimurium) strains were classified. EfficientNetV2 served as the backbone architecture, leveraging fine-grained feature extraction for small targets. Few-shot learning enabled scalability, with domain-adversarial neural networks (DANNs) addressing single domains and multi-DANNs (MDANNs) generalizing across all target domains. The model was trained on source domain data collected under controlled conditions (phase contrast microscopy, 60x magnification, 3-h bacterial incubation) and evaluated on target domains with variations in microscopy modality (brightfield, BF), magnification (20x), and extended incubation to compensate for lower resolution (20x-5h). DANNs improved target domain classification accuracy by up to 54.45% (20x), 43.44% (20x-5h), and 31.67% (BF), with minimal source domain degradation (<4.44%). MDANNs achieved superior performance in the BF domain and substantial gains in the 20x domain. Grad-CAM and t-SNE visualizations validated the model's ability to learn domain-invariant features across diverse conditions. This study presents a scalable and adaptable framework for bacterial classification, reducing reliance on extensive sample preparation and enabling application in decentralized and resource-limited environments.
Spectroscopy Approaches for Food Safety Applications: Improving Data Efficiency Using Active Learning and Semi-Supervised Learning
Oct 24, 2021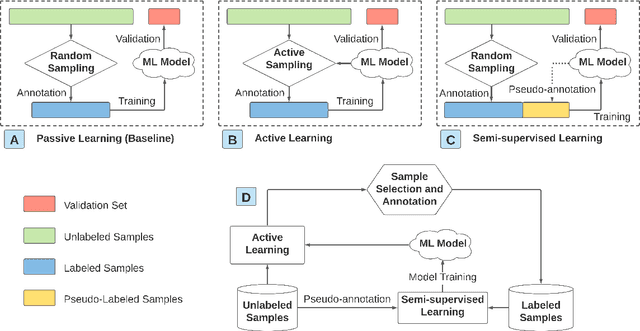
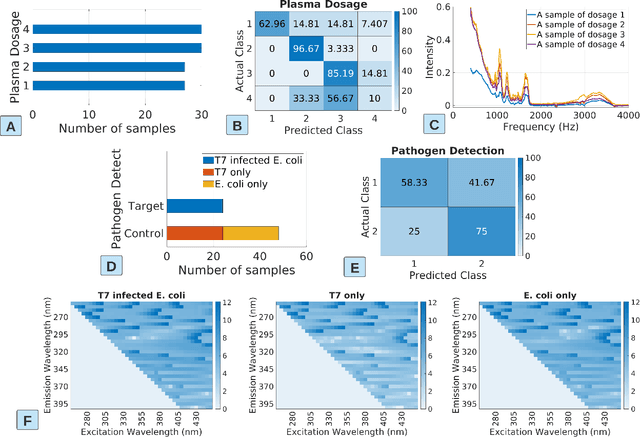
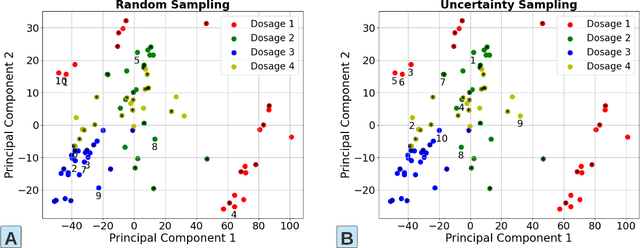
Abstract:The past decade witnesses a rapid development in the measurement and monitoring technologies for food science. Among these technologies, spectroscopy has been widely used for the analysis of food quality, safety, and nutritional properties. Due to the complexity of food systems and the lack of comprehensive predictive models, rapid and simple measurements to predict complex properties in food systems are largely missing. Machine Learning (ML) has shown great potential to improve classification and prediction of these properties. However, the barriers to collect large datasets for ML applications still persists. In this paper, we explore different approaches of data annotation and model training to improve data efficiency for ML applications. Specifically, we leverage Active Learning (AL) and Semi-Supervised Learning (SSL) and investigate four approaches: baseline passive learning, AL, SSL, and a hybrid of AL and SSL. To evaluate these approaches, we collect two spectroscopy datasets: predicting plasma dosage and detecting foodborne pathogen. Our experimental results show that, compared to the de facto passive learning approach, AL and SSL methods reduce the number of labeled samples by 50% and 25% for each ML application, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge