Nina van Tiel
kabr-tools: Automated Framework for Multi-Species Behavioral Monitoring
Oct 02, 2025Abstract:A comprehensive understanding of animal behavior ecology depends on scalable approaches to quantify and interpret complex, multidimensional behavioral patterns. Traditional field observations are often limited in scope, time-consuming, and labor-intensive, hindering the assessment of behavioral responses across landscapes. To address this, we present kabr-tools (Kenyan Animal Behavior Recognition Tools), an open-source package for automated multi-species behavioral monitoring. This framework integrates drone-based video with machine learning systems to extract behavioral, social, and spatial metrics from wildlife footage. Our pipeline leverages object detection, tracking, and behavioral classification systems to generate key metrics, including time budgets, behavioral transitions, social interactions, habitat associations, and group composition dynamics. Compared to ground-based methods, drone-based observations significantly improved behavioral granularity, reducing visibility loss by 15% and capturing more transitions with higher accuracy and continuity. We validate kabr-tools through three case studies, analyzing 969 behavioral sequences, surpassing the capacity of traditional methods for data capture and annotation. We found that, like Plains zebras, vigilance in Grevy's zebras decreases with herd size, but, unlike Plains zebras, habitat has a negligible impact. Plains and Grevy's zebras exhibit strong behavioral inertia, with rare transitions to alert behaviors and observed spatial segregation between Grevy's zebras, Plains zebras, and giraffes in mixed-species herds. By enabling automated behavioral monitoring at scale, kabr-tools offers a powerful tool for ecosystem-wide studies, advancing conservation, biodiversity research, and ecological monitoring.
MaskSDM with Shapley values to improve flexibility, robustness, and explainability in species distribution modeling
Mar 17, 2025Abstract:Species Distribution Models (SDMs) play a vital role in biodiversity research, conservation planning, and ecological niche modeling by predicting species distributions based on environmental conditions. The selection of predictors is crucial, strongly impacting both model accuracy and how well the predictions reflect ecological patterns. To ensure meaningful insights, input variables must be carefully chosen to match the study objectives and the ecological requirements of the target species. However, existing SDMs, including both traditional and deep learning-based approaches, often lack key capabilities for variable selection: (i) flexibility to choose relevant predictors at inference without retraining; (ii) robustness to handle missing predictor values without compromising accuracy; and (iii) explainability to interpret and accurately quantify each predictor's contribution. To overcome these limitations, we introduce MaskSDM, a novel deep learning-based SDM that enables flexible predictor selection by employing a masked training strategy. This approach allows the model to make predictions with arbitrary subsets of input variables while remaining robust to missing data. It also provides a clearer understanding of how adding or removing a given predictor affects model performance and predictions. Additionally, MaskSDM leverages Shapley values for precise predictor contribution assessments, improving upon traditional approximations. We evaluate MaskSDM on the global sPlotOpen dataset, modeling the distributions of 12,738 plant species. Our results show that MaskSDM outperforms imputation-based methods and approximates models trained on specific subsets of variables. These findings underscore MaskSDM's potential to increase the applicability and adoption of SDMs, laying the groundwork for developing foundation models in SDMs that can be readily applied to diverse ecological applications.
Multi-Scale and Multimodal Species Distribution Modeling
Nov 06, 2024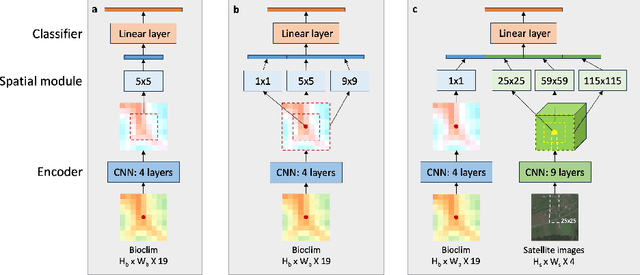
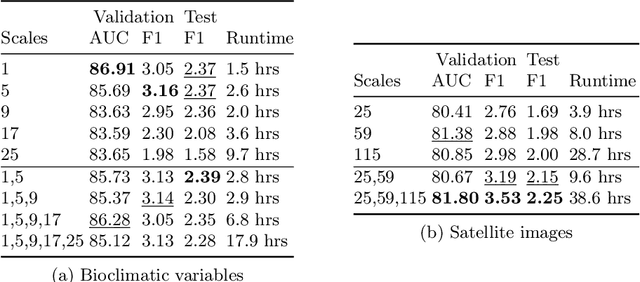
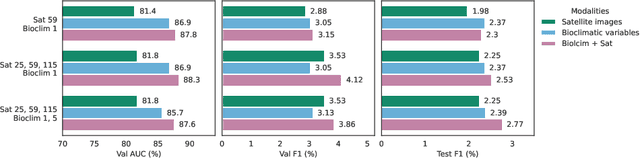
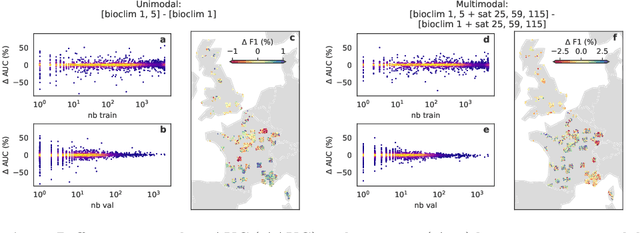
Abstract:Species distribution models (SDMs) aim to predict the distribution of species by relating occurrence data with environmental variables. Recent applications of deep learning to SDMs have enabled new avenues, specifically the inclusion of spatial data (environmental rasters, satellite images) as model predictors, allowing the model to consider the spatial context around each species' observations. However, the appropriate spatial extent of the images is not straightforward to determine and may affect the performance of the model, as scale is recognized as an important factor in SDMs. We develop a modular structure for SDMs that allows us to test the effect of scale in both single- and multi-scale settings. Furthermore, our model enables different scales to be considered for different modalities, using a late fusion approach. Results on the GeoLifeCLEF 2023 benchmark indicate that considering multimodal data and learning multi-scale representations leads to more accurate models.
Imbalance-aware Presence-only Loss Function for Species Distribution Modeling
Mar 12, 2024



Abstract:In the face of significant biodiversity decline, species distribution models (SDMs) are essential for understanding the impact of climate change on species habitats by connecting environmental conditions to species occurrences. Traditionally limited by a scarcity of species observations, these models have significantly improved in performance through the integration of larger datasets provided by citizen science initiatives. However, they still suffer from the strong class imbalance between species within these datasets, often resulting in the penalization of rare species--those most critical for conservation efforts. To tackle this issue, this study assesses the effectiveness of training deep learning models using a balanced presence-only loss function on large citizen science-based datasets. We demonstrate that this imbalance-aware loss function outperforms traditional loss functions across various datasets and tasks, particularly in accurately modeling rare species with limited observations.
On the selection and effectiveness of pseudo-absences for species distribution modeling with deep learning
Jan 03, 2024



Abstract:Species distribution modeling is a highly versatile tool for understanding the intricate relationship between environmental conditions and species occurrences. However, the available data often lacks information on confirmed species absence and is limited to opportunistically sampled, presence-only observations. To overcome this limitation, a common approach is to employ pseudo-absences, which are specific geographic locations designated as negative samples. While pseudo-absences are well-established for single-species distribution models, their application in the context of multi-species neural networks remains underexplored. Notably, the significant class imbalance between species presences and pseudo-absences is often left unaddressed. Moreover, the existence of different types of pseudo-absences (e.g., random and target-group background points) adds complexity to the selection process. Determining the optimal combination of pseudo-absences types is difficult and depends on the characteristics of the data, particularly considering that certain types of pseudo-absences can be used to mitigate geographic biases. In this paper, we demonstrate that these challenges can be effectively tackled by integrating pseudo-absences in the training of multi-species neural networks through modifications to the loss function. This adjustment involves assigning different weights to the distinct terms of the loss function, thereby addressing both the class imbalance and the choice of pseudo-absence types. Additionally, we propose a strategy to set these loss weights using spatial block cross-validation with presence-only data. We evaluate our approach using a benchmark dataset containing independent presence-absence data from six different regions and report improved results when compared to competing approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge