Naveen Himthani
CLAIRE -- Parallelized Diffeomorphic Image Registration for Large-Scale Biomedical Imaging Applications
Sep 16, 2022



Abstract:We study the performance of CLAIRE -- a diffeomorphic multi-node, multi-GPU image-registration algorithm, and software -- in large-scale biomedical imaging applications with billions of voxels. At such resolutions, most existing software packages for diffeomorphic image registration are prohibitively expensive. As a result, practitioners first significantly downsample the original images and then register them using existing tools. Our main contribution is an extensive analysis of the impact of downsampling on registration performance. We study this impact by comparing full-resolution registrations obtained with CLAIRE to lower-resolution registrations for synthetic and real-world imaging datasets. Our results suggest that registration at full resolution can yield a superior registration quality -- but not always. For example, downsampling a synthetic image from $1024^3$ to $256^3$ decreases the Dice coefficient from 92% to 79%. However, the differences are less pronounced for noisy or low-contrast high-resolution images. CLAIRE allows us not only to register images of clinically relevant size in a few seconds but also to register images at unprecedented resolution in a reasonable time. The highest resolution considered is CLARITY images of size $2816\times3016\times1162$. To the best of our knowledge, this is the first study on image registration quality at such resolutions.
A Novel Domain Adaptation Framework for Medical Image Segmentation
Oct 11, 2018

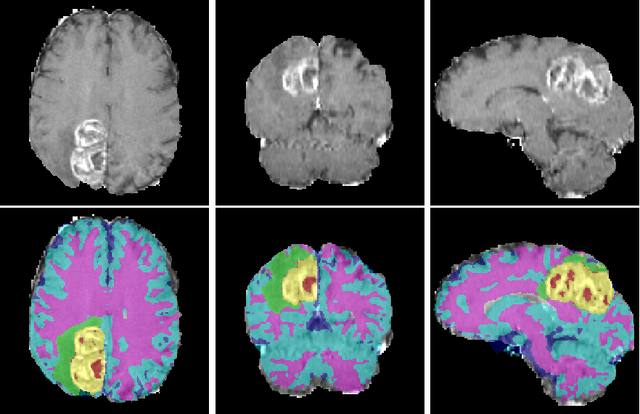

Abstract:We propose a segmentation framework that uses deep neural networks and introduce two innovations. First, we describe a biophysics-based domain adaptation method. Second, we propose an automatic method to segment white and gray matter, and cerebrospinal fluid, in addition to tumorous tissue. Regarding our first innovation, we use a domain adaptation framework that combines a novel multispecies biophysical tumor growth model with a generative adversarial model to create realistic looking synthetic multimodal MR images with known segmentation. Regarding our second innovation, we propose an automatic approach to enrich available segmentation data by computing the segmentation for healthy tissues. This segmentation, which is done using diffeomorphic image registration between the BraTS training data and a set of prelabeled atlases, provides more information for training and reduces the class imbalance problem. Our overall approach is not specific to any particular neural network and can be used in conjunction with existing solutions. We demonstrate the performance improvement using a 2D U-Net for the BraTS'18 segmentation challenge. Our biophysics based domain adaptation achieves better results, as compared to the existing state-of-the-art GAN model used to create synthetic data for training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge