Nathaniel Diamant
Conformalized Deep Splines for Optimal and Efficient Prediction Sets
Nov 01, 2023



Abstract:Uncertainty estimation is critical in high-stakes machine learning applications. One effective way to estimate uncertainty is conformal prediction, which can provide predictive inference with statistical coverage guarantees. We present a new conformal regression method, Spline Prediction Intervals via Conformal Estimation (SPICE), that estimates the conditional density using neural-network-parameterized splines. We prove universal approximation and optimality results for SPICE, which are empirically validated by our experiments. SPICE is compatible with two different efficient-to-compute conformal scores, one oracle-optimal for marginal coverage (SPICE-ND) and the other asymptotically optimal for conditional coverage (SPICE-HPD). Results on benchmark datasets demonstrate SPICE-ND models achieve the smallest average prediction set sizes, including average size reductions of nearly 50% for some datasets compared to the next best baseline. SPICE-HPD models achieve the best conditional coverage compared to baselines. The SPICE implementation is made available.
Complex Preferences for Different Convergent Priors in Discrete Graph Diffusion
Jun 21, 2023
Abstract:Diffusion models have achieved state-of-the-art performance in generating many different kinds of data, including images, text, and videos. Despite their success, there has been limited research on how the underlying diffusion process and the final convergent prior can affect generative performance; this research has also been limited to continuous data types and a score-based diffusion framework. To fill this gap, we explore how different discrete diffusion kernels (which converge to different prior distributions) affect the performance of diffusion models for graphs. To this end, we developed a novel formulation of a family of discrete diffusion kernels which are easily adjustable to converge to different Bernoulli priors, and we study the effect of these different kernels on generative performance. We show that the quality of generated graphs is sensitive to the prior used, and that the optimal choice cannot be explained by obvious statistics or metrics, which challenges the intuitions which previous works have suggested.
GraphGUIDE: interpretable and controllable conditional graph generation with discrete Bernoulli diffusion
Feb 07, 2023



Abstract:Diffusion models achieve state-of-the-art performance in generating realistic objects and have been successfully applied to images, text, and videos. Recent work has shown that diffusion can also be defined on graphs, including graph representations of drug-like molecules. Unfortunately, it remains difficult to perform conditional generation on graphs in a way which is interpretable and controllable. In this work, we propose GraphGUIDE, a novel framework for graph generation using diffusion models, where edges in the graph are flipped or set at each discrete time step. We demonstrate GraphGUIDE on several graph datasets, and show that it enables full control over the conditional generation of arbitrary structural properties without relying on predefined labels. Our framework for graph diffusion can have a large impact on the interpretable conditional generation of graphs, including the generation of drug-like molecules with desired properties in a way which is informed by experimental evidence.
Improving Graph Generation by Restricting Graph Bandwidth
Jan 25, 2023



Abstract:Deep graph generative modeling has proven capable of learning the distribution of complex, multi-scale structures characterizing real-world graphs. However, one of the main limitations of existing methods is their large output space, which limits generation scalability and hinders accurate modeling of the underlying distribution. To overcome these limitations, we propose a novel approach that significantly reduces the output space of existing graph generative models. Specifically, starting from the observation that many real-world graphs have low graph bandwidth, we restrict graph bandwidth during training and generation. Our strategy improves both generation scalability and quality without increasing architectural complexity or reducing expressiveness. Our approach is compatible with existing graph generative methods, and we describe its application to both autoregressive and one-shot models. We extensively validate our strategy on synthetic and real datasets, including molecular graphs. Our experiments show that, in addition to improving generation efficiency, our approach consistently improves generation quality and reconstruction accuracy. The implementation is made available.
Patient Contrastive Learning: a Performant, Expressive, and Practical Approach to ECG Modeling
Apr 09, 2021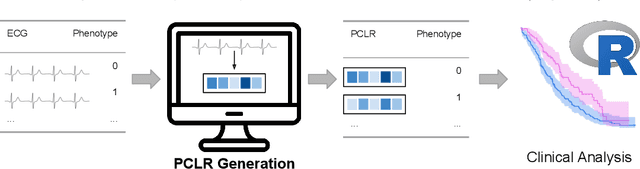
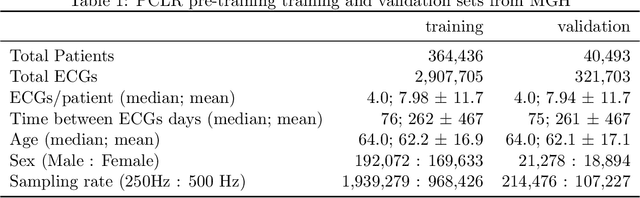
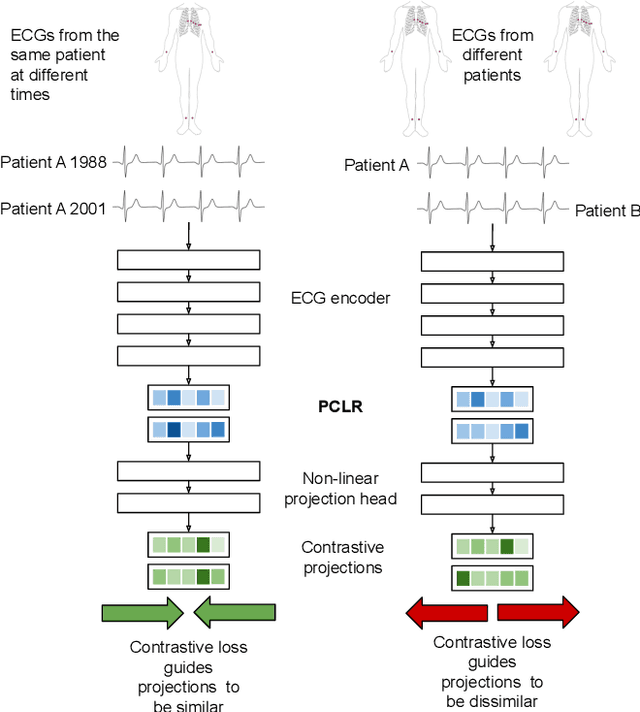
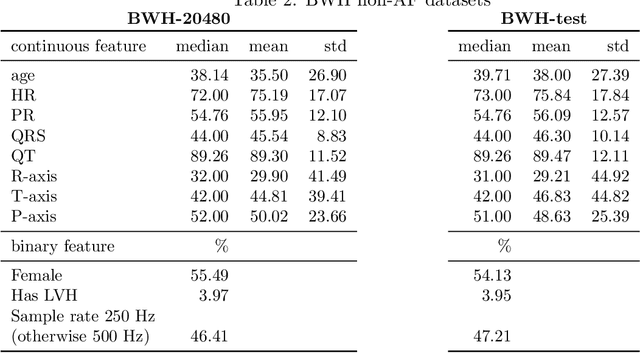
Abstract:Supervised machine learning applications in health care are often limited due to a scarcity of labeled training data. To mitigate this effect of small sample size, we introduce a pre-training approach, Patient Contrastive Learning of Representations (PCLR), which creates latent representations of ECGs from a large number of unlabeled examples. The resulting representations are expressive, performant, and practical across a wide spectrum of clinical tasks. We develop PCLR using a large health care system with over 3.2 million 12-lead ECGs, and demonstrate substantial improvements across multiple new tasks when there are fewer than 5,000 labels. We release our model to extract ECG representations at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/PCLR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge