Erik Reinertsen
The Patient is not a Moving Document: A World Model Training Paradigm for Longitudinal EHR
Jan 29, 2026Abstract:Large language models (LLMs) trained with next-word-prediction have achieved success as clinical foundation models. Representations from these language backbones yield strong linear probe performance across biomedical tasks, suggesting that patient semantics emerge from next-token prediction at scale. However, this paradigm treats patients as a document to be summarized rather than a dynamical system to be simulated; a patient's trajectory emerges from their state evolving under interventions and time, requiring models that simulate dynamics rather than predict tokens. To address this, we introduce SMB-Structure, a world model for structured EHR that grounds a joint-embedding prediction architecture (JEPA) with next-token prediction (SFT). SFT grounds our model to reconstruct future patient states in token space, while JEPA predicts those futures in latent space from the initial patient representation alone, forcing trajectory dynamics to be encoded before the next state is observed. We validate across two large-scale cohorts: Memorial Sloan Kettering (23,319 oncology patients; 323,000+ patient-years) and INSPECT (19,402 pulmonary embolism patients). Using a linear probe evaluated at multiple points along the disease trajectory, we demonstrate that our training paradigm learns embeddings that capture disease dynamics not recoverable by autoregressive baselines, enabling SMB-Structure to achieve competitive performance on complex tasks characterized by high patient heterogeneity. Model weights are available at https://huggingface.co/standardmodelbio/SMB-v1-1.7B-Structure.
Benchmarking changepoint detection algorithms on cardiac time series
Apr 16, 2024
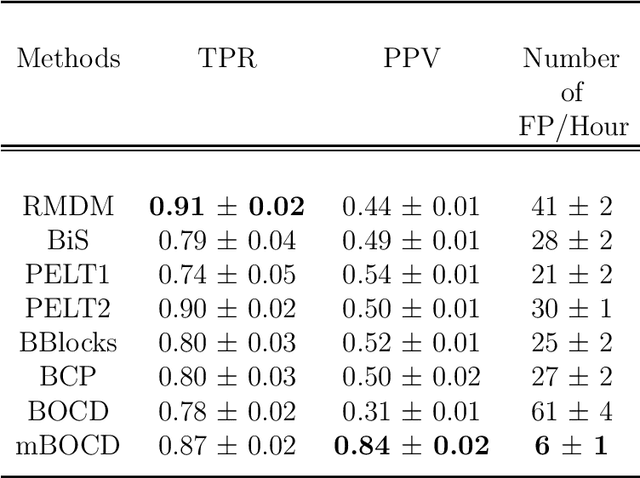

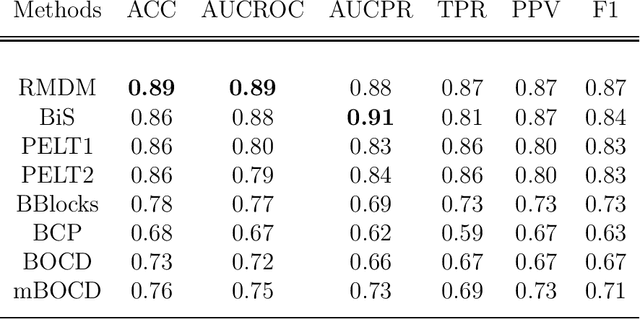
Abstract:The pattern of state changes in a biomedical time series can be related to health or disease. This work presents a principled approach for selecting a changepoint detection algorithm for a specific task, such as disease classification. Eight key algorithms were compared, and the performance of each algorithm was evaluated as a function of temporal tolerance, noise, and abnormal conduction (ectopy) on realistic artificial cardiovascular time series data. All algorithms were applied to real data (cardiac time series of 22 patients with REM-behavior disorder (RBD) and 15 healthy controls) using the parameters selected on artificial data. Finally, features were derived from the detected changepoints to classify RBD patients from healthy controls using a K-Nearest Neighbors approach. On artificial data, Modified Bayesian Changepoint Detection algorithm provided superior positive predictive value for state change identification while Recursive Mean Difference Maximization (RMDM) achieved the highest true positive rate. For the classification task, features derived from the RMDM algorithm provided the highest leave one out cross validated accuracy of 0.89 and true positive rate of 0.87. Automatically detected changepoints provide useful information about subject's physiological state which cannot be directly observed. However, the choice of change point detection algorithm depends on the nature of the underlying data and the downstream application, such as a classification task. This work represents the first time change point detection algorithms have been compared in a meaningful way and utilized in a classification task, which demonstrates the effect of changepoint algorithm choice on application performance.
Patient Contrastive Learning: a Performant, Expressive, and Practical Approach to ECG Modeling
Apr 09, 2021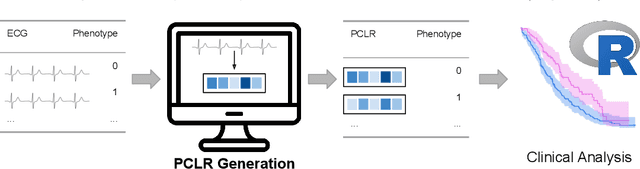
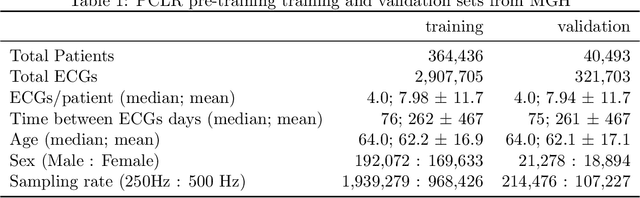
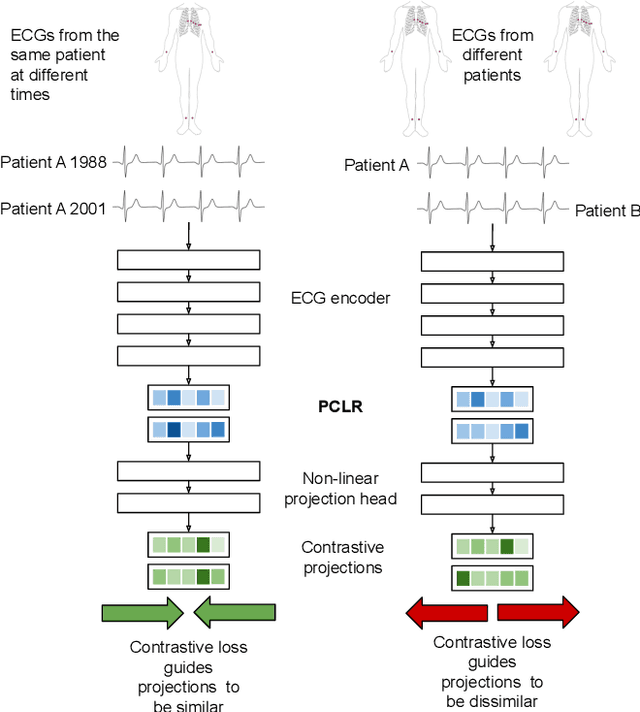
Abstract:Supervised machine learning applications in health care are often limited due to a scarcity of labeled training data. To mitigate this effect of small sample size, we introduce a pre-training approach, Patient Contrastive Learning of Representations (PCLR), which creates latent representations of ECGs from a large number of unlabeled examples. The resulting representations are expressive, performant, and practical across a wide spectrum of clinical tasks. We develop PCLR using a large health care system with over 3.2 million 12-lead ECGs, and demonstrate substantial improvements across multiple new tasks when there are fewer than 5,000 labels. We release our model to extract ECG representations at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/PCLR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge