Ayse Cakmak
Benchmarking changepoint detection algorithms on cardiac time series
Apr 16, 2024
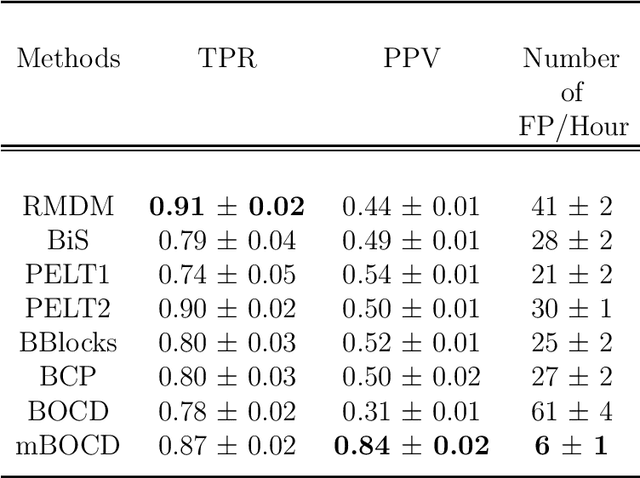

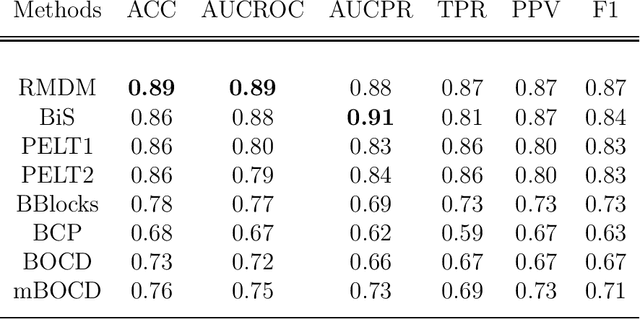
Abstract:The pattern of state changes in a biomedical time series can be related to health or disease. This work presents a principled approach for selecting a changepoint detection algorithm for a specific task, such as disease classification. Eight key algorithms were compared, and the performance of each algorithm was evaluated as a function of temporal tolerance, noise, and abnormal conduction (ectopy) on realistic artificial cardiovascular time series data. All algorithms were applied to real data (cardiac time series of 22 patients with REM-behavior disorder (RBD) and 15 healthy controls) using the parameters selected on artificial data. Finally, features were derived from the detected changepoints to classify RBD patients from healthy controls using a K-Nearest Neighbors approach. On artificial data, Modified Bayesian Changepoint Detection algorithm provided superior positive predictive value for state change identification while Recursive Mean Difference Maximization (RMDM) achieved the highest true positive rate. For the classification task, features derived from the RMDM algorithm provided the highest leave one out cross validated accuracy of 0.89 and true positive rate of 0.87. Automatically detected changepoints provide useful information about subject's physiological state which cannot be directly observed. However, the choice of change point detection algorithm depends on the nature of the underlying data and the downstream application, such as a classification task. This work represents the first time change point detection algorithms have been compared in a meaningful way and utilized in a classification task, which demonstrates the effect of changepoint algorithm choice on application performance.
Addressing Class Imbalance in Classification Problems of Noisy Signals by using Fourier Transform Surrogates
Jun 20, 2018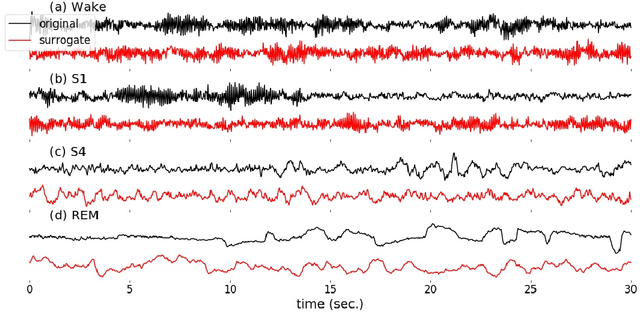
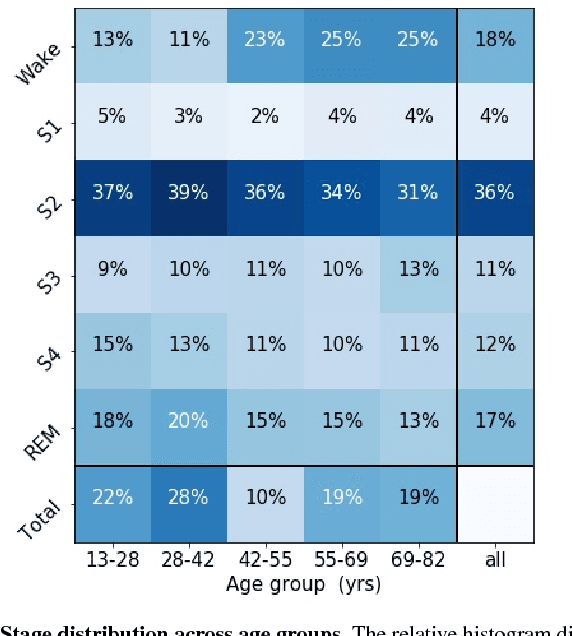
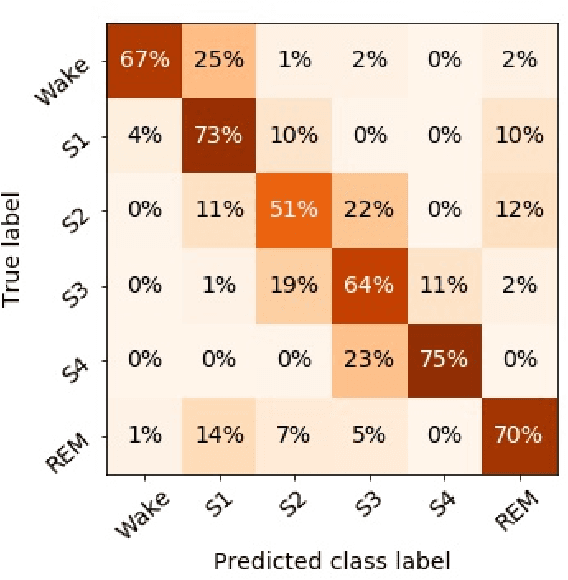
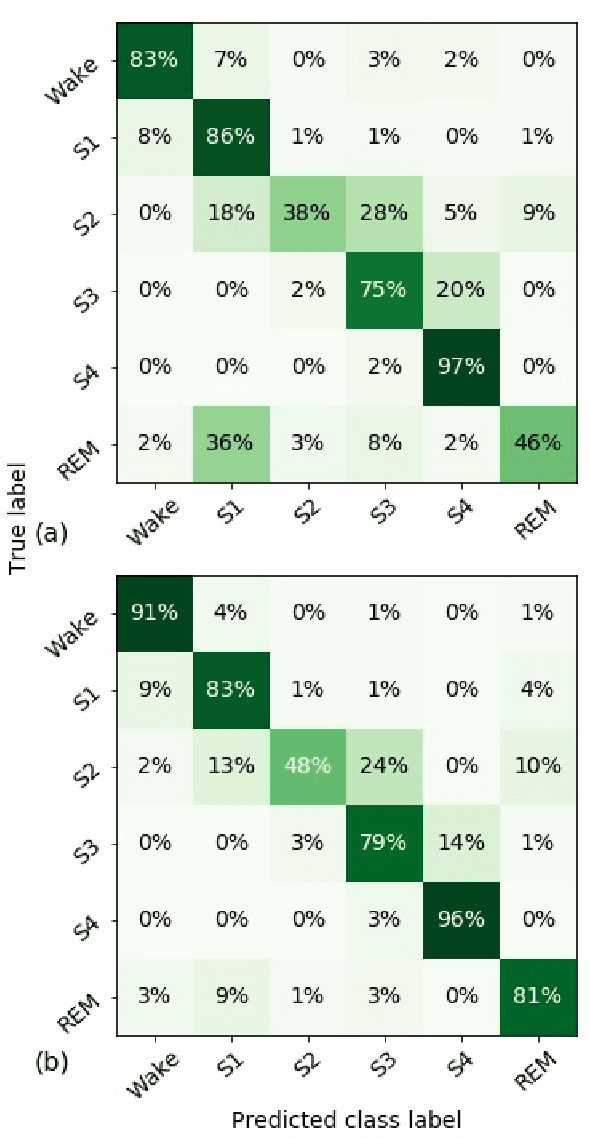
Abstract:Randomizing the Fourier-transform (FT) phases of temporal-spatial data generates surrogates that approximate examples from the data-generating distribution. We propose such FT surrogates as a novel tool to augment and analyze training of neural networks and explore the approach in the example of sleep-stage classification. By computing FT surrogates of raw EEG, EOG, and EMG signals of under-represented sleep stages, we balanced the CAPSLPDB sleep database. We then trained and tested a convolutional neural network for sleep stage classification, and found that our surrogate-based augmentation improved the mean F1-score by 7%. As another application of FT surrogates, we formulated an approach to compute saliency maps for individual sleep epochs. The visualization is based on the response of inferred class probabilities under replacement of short data segments by partial surrogates. To quantify how well the distributions of the surrogates and the original data match, we evaluated a trained classifier on surrogates of correctly classified examples, and summarized these conditional predictions in a confusion matrix. We show how such conditional confusion matrices can qualitatively explain the performance of surrogates in class balancing. The FT-surrogate augmentation approach may improve classification on noisy signals if carefully adapted to the data distribution under analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge