Justus T. C. Schwabedal
SUMO: Advanced sleep spindle identification with neural networks
Feb 06, 2022



Abstract:Sleep spindles are neurophysiological phenomena that appear to be linked to memory formation and other functions of the central nervous system, and that can be observed in electroencephalographic recordings (EEG) during sleep. Manually identified spindle annotations in EEG recordings suffer from substantial intra- and inter-rater variability, even if raters have been highly trained, which reduces the reliability of spindle measures as a research and diagnostic tool. The Massive Online Data Annotation (MODA) project has recently addressed this problem by forming a consensus from multiple such rating experts, thus providing a corpus of spindle annotations of enhanced quality. Based on this dataset, we present a U-Net-type deep neural network model to automatically detect sleep spindles. Our model's performance exceeds that of the state-of-the-art detector and of most experts in the MODA dataset. We observed improved detection accuracy in subjects of all ages, including older individuals whose spindles are particularly challenging to detect reliably. Our results underline the potential of automated methods to do repetitive cumbersome tasks with super-human performance.
Automated scoring of pre-REM sleep in mice with deep learning
May 05, 2021



Abstract:Reliable automation of the labor-intensive manual task of scoring animal sleep can facilitate the analysis of long-term sleep studies. In recent years, deep-learning-based systems, which learn optimal features from the data, increased scoring accuracies for the classical sleep stages of Wake, REM, and Non-REM. Meanwhile, it has been recognized that the statistics of transitional stages such as pre-REM, found between Non-REM and REM, may hold additional insight into the physiology of sleep and are now under vivid investigation. We propose a classification system based on a simple neural network architecture that scores the classical stages as well as pre-REM sleep in mice. When restricted to the classical stages, the optimized network showed state-of-the-art classification performance with an out-of-sample F1 score of 0.95. When unrestricted, the network showed lower F1 scores on pre-REM (0.5) compared to the classical stages. The result is comparable to previous attempts to score transitional stages in other species such as transition sleep in rats or N1 sleep in humans. Nevertheless, we observed that the sequence of predictions including pre-REM typically transitioned from Non-REM to REM reflecting sleep dynamics observed by human scorers. Our findings provide further evidence for the difficulty of scoring transitional sleep stages, likely because such stages of sleep are under-represented in typical data sets or show large inter-scorer variability. We further provide our source code and an online platform to run predictions with our trained network.
Differentially Private Generation of Small Images
May 06, 2020


Abstract:We explore the training of generative adversarial networks with differential privacy to anonymize image data sets. On MNIST, we numerically measure the privacy-utility trade-off using parameters from $\epsilon$-$\delta$ differential privacy and the inception score. Our experiments uncover a saturated training regime where an increasing privacy budget adds little to the quality of generated images. We also explain analytically why differentially private Adam optimization is independent of the gradient clipping parameter. Furthermore, we highlight common errors in previous works on differentially private deep learning, which we uncovered in recent literature. Throughout the treatment of the subject, we hope to prevent erroneous estimates of anonymity in the future.
Automated Classification of Sleep Stages and EEG Artifacts in Mice with Deep Learning
Sep 22, 2018



Abstract:Sleep scoring is a necessary and time-consuming task in sleep studies. In animal models (such as mice) or in humans, automating this tedious process promises to facilitate long-term studies and to promote sleep biology as a data-driven field. We introduce a deep neural network model that is able to predict different states of consciousness (Wake, Non-REM, REM) in mice from EEG and EMG recordings with excellent scoring results for out-of-sample data. Predictions are made on epochs of 4 seconds length, and epochs are classified as artifact-free or not. The model architecture draws on recent advances in deep learning and in convolutional neural networks research. In contrast to previous approaches towards automated sleep scoring, our model does not rely on manually defined features of the data but learns predictive features automatically. We expect deep learning models like ours to become widely applied in different fields, automating many repetitive cognitive tasks that were previously difficult to tackle.
Addressing Class Imbalance in Classification Problems of Noisy Signals by using Fourier Transform Surrogates
Jun 20, 2018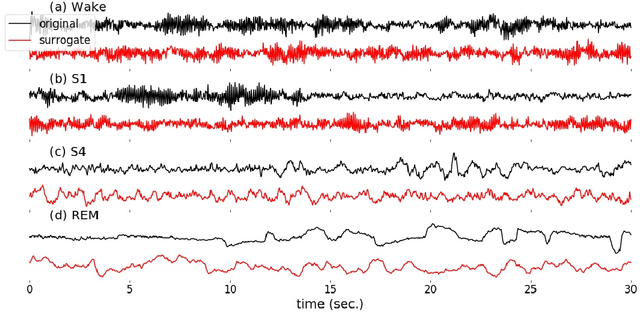
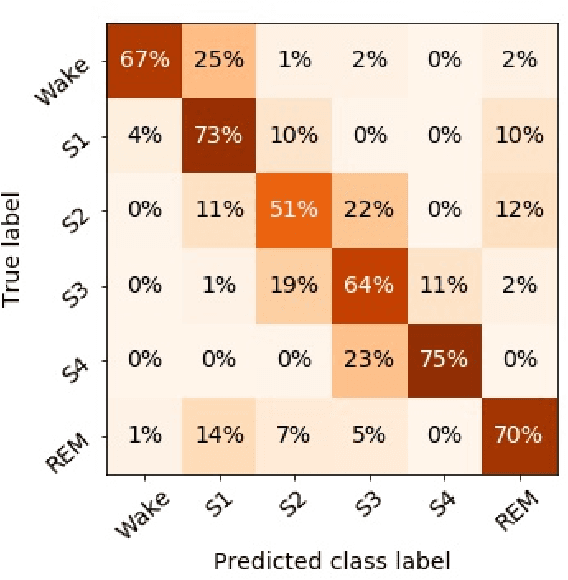
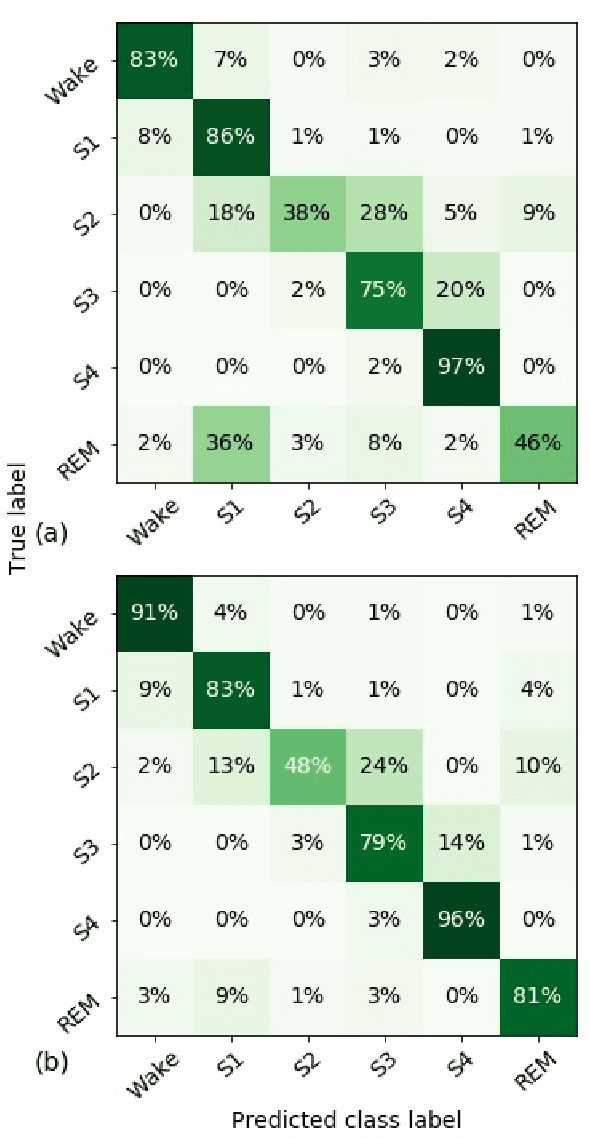
Abstract:Randomizing the Fourier-transform (FT) phases of temporal-spatial data generates surrogates that approximate examples from the data-generating distribution. We propose such FT surrogates as a novel tool to augment and analyze training of neural networks and explore the approach in the example of sleep-stage classification. By computing FT surrogates of raw EEG, EOG, and EMG signals of under-represented sleep stages, we balanced the CAPSLPDB sleep database. We then trained and tested a convolutional neural network for sleep stage classification, and found that our surrogate-based augmentation improved the mean F1-score by 7%. As another application of FT surrogates, we formulated an approach to compute saliency maps for individual sleep epochs. The visualization is based on the response of inferred class probabilities under replacement of short data segments by partial surrogates. To quantify how well the distributions of the surrogates and the original data match, we evaluated a trained classifier on surrogates of correctly classified examples, and summarized these conditional predictions in a confusion matrix. We show how such conditional confusion matrices can qualitatively explain the performance of surrogates in class balancing. The FT-surrogate augmentation approach may improve classification on noisy signals if carefully adapted to the data distribution under analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge