Nathan Wan
Bridging the Generalization Gap: Training Robust Models on Confounded Biological Data
Dec 12, 2018



Abstract:Statistical learning on biological data can be challenging due to confounding variables in sample collection and processing. Confounders can cause models to generalize poorly and result in inaccurate prediction performance metrics if models are not validated thoroughly. In this paper, we propose methods to control for confounding factors and further improve prediction performance. We introduce OrthoNormal basis construction In cOnfounding factor Normalization (ONION) to remove confounding covariates and use the Domain-Adversarial Neural Network (DANN) to penalize models for encoding confounder information. We apply the proposed methods to simulated and empirical patient data and show significant improvements in generalization.
METCC: METric learning for Confounder Control Making distance matter in high dimensional biological analysis
Dec 07, 2018



Abstract:High-dimensional data acquired from biological experiments such as next generation sequencing are subject to a number of confounding effects. These effects include both technical effects, such as variation across batches from instrument noise or sample processing, or institution-specific differences in sample acquisition and physical handling, as well as biological effects arising from true but irrelevant differences in the biology of each sample, such as age biases in diseases. Prior work has used linear methods to adjust for such batch effects. Here, we apply contrastive metric learning by a non-linear triplet network to optimize the ability to distinguish biologically distinct sample classes in the presence of irrelevant technical and biological variation. Using whole-genome cell-free DNA data from 817 patients, we demonstrate that our approach, METric learning for Confounder Control (METCC), is able to match or exceed the classification performance achieved using a best-in-class linear method (HCP) or no normalization. Critically, results from METCC appear less confounded by irrelevant technical variables like institution and batch than those from other methods even without access to high quality metadata information required by many existing techniques; offering hope for improved generalization.
Speech recognition for medical conversations
Jun 20, 2018
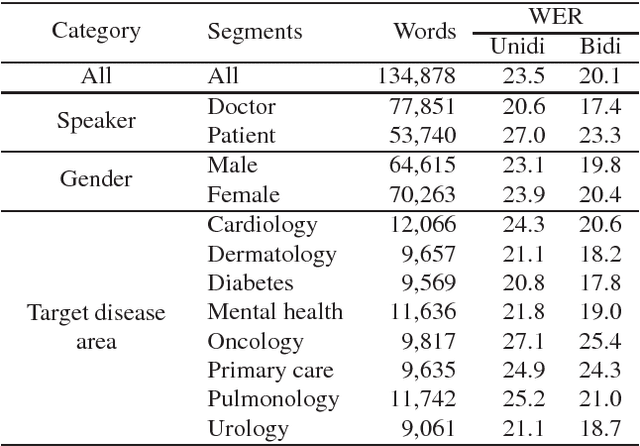
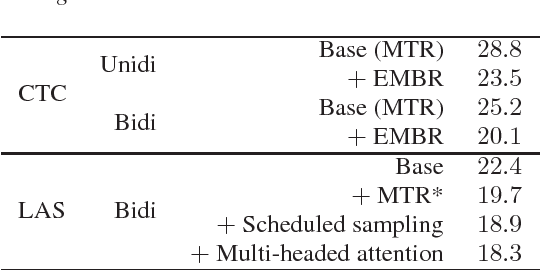
Abstract:In this work we explored building automatic speech recognition models for transcribing doctor patient conversation. We collected a large scale dataset of clinical conversations ($14,000$ hr), designed the task to represent the real word scenario, and explored several alignment approaches to iteratively improve data quality. We explored both CTC and LAS systems for building speech recognition models. The LAS was more resilient to noisy data and CTC required more data clean up. A detailed analysis is provided for understanding the performance for clinical tasks. Our analysis showed the speech recognition models performed well on important medical utterances, while errors occurred in causal conversations. Overall we believe the resulting models can provide reasonable quality in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge