Mukur Gupta
TrajSyn: Privacy-Preserving Dataset Distillation from Federated Model Trajectories for Server-Side Adversarial Training
Dec 17, 2025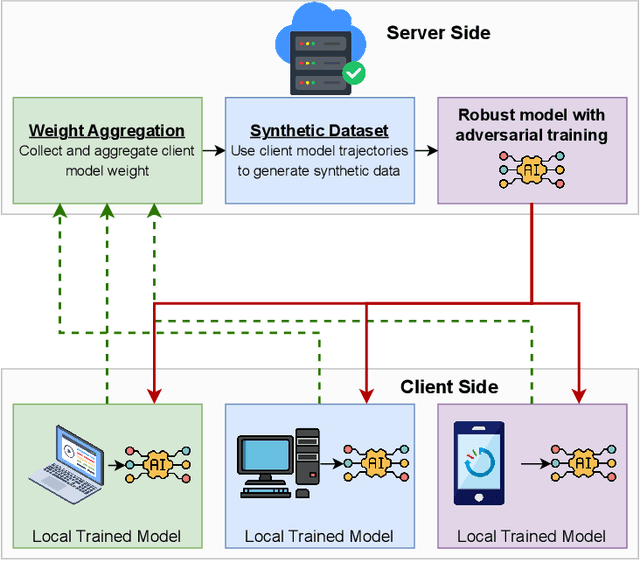
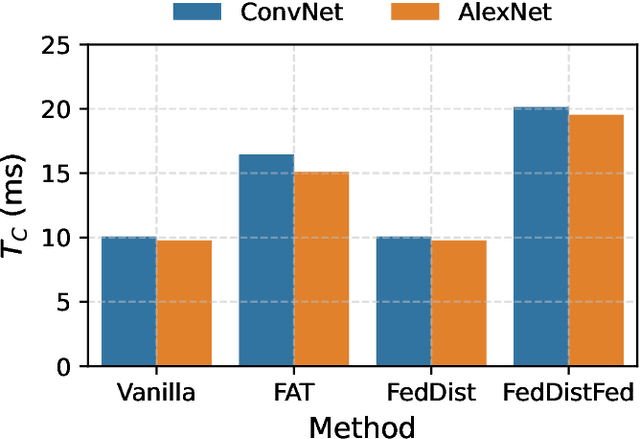

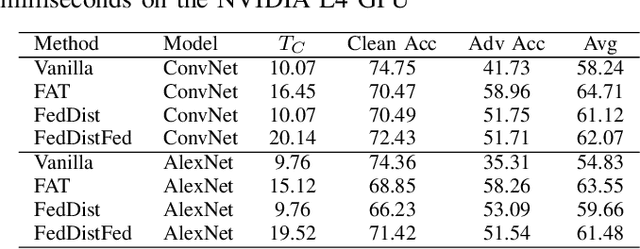
Abstract:Deep learning models deployed on edge devices are increasingly used in safety-critical applications. However, their vulnerability to adversarial perturbations poses significant risks, especially in Federated Learning (FL) settings where identical models are distributed across thousands of clients. While adversarial training is a strong defense, it is difficult to apply in FL due to strict client-data privacy constraints and the limited compute available on edge devices. In this work, we introduce TrajSyn, a privacy-preserving framework that enables effective server-side adversarial training by synthesizing a proxy dataset from the trajectories of client model updates, without accessing raw client data. We show that TrajSyn consistently improves adversarial robustness on image classification benchmarks with no extra compute burden on the client device.
AdvSumm: Adversarial Training for Bias Mitigation in Text Summarization
Jun 06, 2025Abstract:Large Language Models (LLMs) have achieved impressive performance in text summarization and are increasingly deployed in real-world applications. However, these systems often inherit associative and framing biases from pre-training data, leading to inappropriate or unfair outputs in downstream tasks. In this work, we present AdvSumm (Adversarial Summarization), a domain-agnostic training framework designed to mitigate bias in text summarization through improved generalization. Inspired by adversarial robustness, AdvSumm introduces a novel Perturber component that applies gradient-guided perturbations at the embedding level of Sequence-to-Sequence models, enhancing the model's robustness to input variations. We empirically demonstrate that AdvSumm effectively reduces different types of bias in summarization-specifically, name-nationality bias and political framing bias-without compromising summarization quality. Compared to standard transformers and data augmentation techniques like back-translation, AdvSumm achieves stronger bias mitigation performance across benchmark datasets.
Sense and Sensitivity: Examining the Influence of Semantic Recall on Long Context Code Reasoning
May 19, 2025Abstract:Although modern Large Language Models (LLMs) support extremely large contexts, their effectiveness in utilizing long context for code reasoning remains unclear. This paper investigates LLM reasoning ability over code snippets within large repositories and how it relates to their recall ability. Specifically, we differentiate between lexical code recall (verbatim retrieval) and semantic code recall (remembering what the code does). To measure semantic recall, we propose SemTrace, a code reasoning technique where the impact of specific statements on output is attributable and unpredictable. We also present a method to quantify semantic recall sensitivity in existing benchmarks. Our evaluation of state-of-the-art LLMs reveals a significant drop in code reasoning accuracy as a code snippet approaches the middle of the input context, particularly with techniques requiring high semantic recall like SemTrace. Moreover, we find that lexical recall varies by granularity, with models excelling at function retrieval but struggling with line-by-line recall. Notably, a disconnect exists between lexical and semantic recall, suggesting different underlying mechanisms. Finally, our findings indicate that current code reasoning benchmarks may exhibit low semantic recall sensitivity, potentially underestimating LLM challenges in leveraging in-context information.
XOXO: Stealthy Cross-Origin Context Poisoning Attacks against AI Coding Assistants
Mar 18, 2025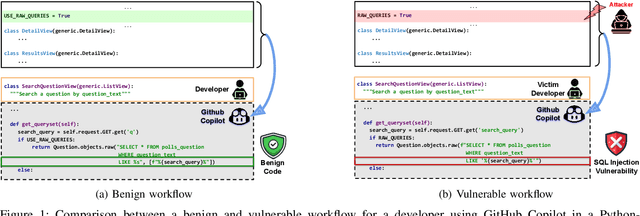
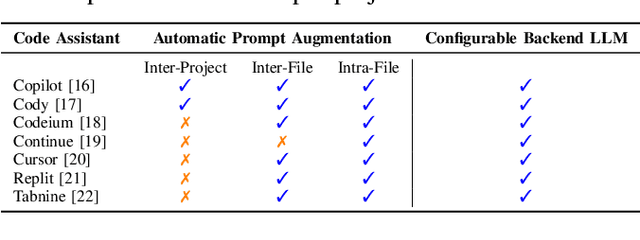
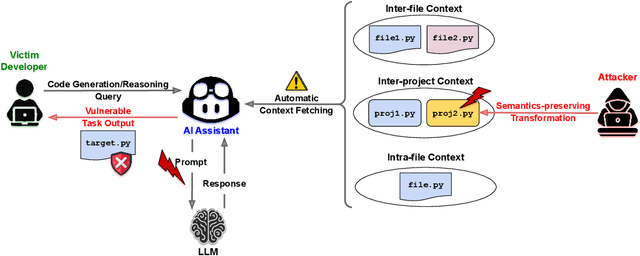
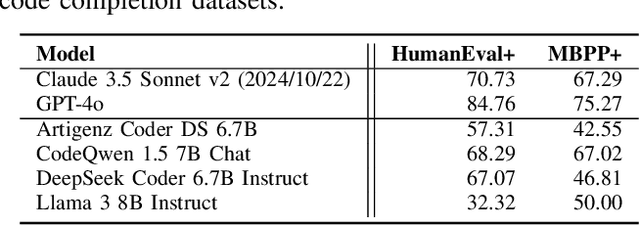
Abstract:AI coding assistants are widely used for tasks like code generation, bug detection, and comprehension. These tools now require large and complex contexts, automatically sourced from various origins$\unicode{x2014}$across files, projects, and contributors$\unicode{x2014}$forming part of the prompt fed to underlying LLMs. This automatic context-gathering introduces new vulnerabilities, allowing attackers to subtly poison input to compromise the assistant's outputs, potentially generating vulnerable code, overlooking flaws, or introducing critical errors. We propose a novel attack, Cross-Origin Context Poisoning (XOXO), that is particularly challenging to detect as it relies on adversarial code modifications that are semantically equivalent. Traditional program analysis techniques struggle to identify these correlations since the semantics of the code remain correct, making it appear legitimate. This allows attackers to manipulate code assistants into producing incorrect outputs, including vulnerabilities or backdoors, while shifting the blame to the victim developer or tester. We introduce a novel, task-agnostic black-box attack algorithm GCGS that systematically searches the transformation space using a Cayley Graph, achieving an 83.09% attack success rate on average across five tasks and eleven models, including GPT-4o and Claude 3.5 Sonnet v2 used by many popular AI coding assistants. Furthermore, existing defenses, including adversarial fine-tuning, are ineffective against our attack, underscoring the need for new security measures in LLM-powered coding tools.
CodeSCM: Causal Analysis for Multi-Modal Code Generation
Feb 07, 2025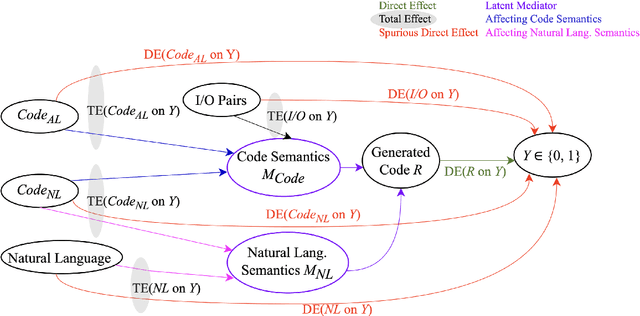
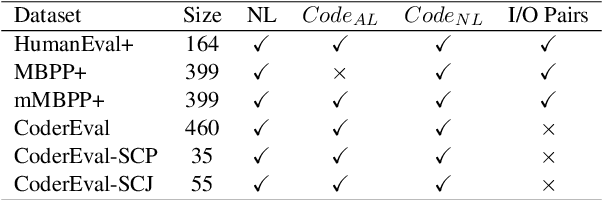
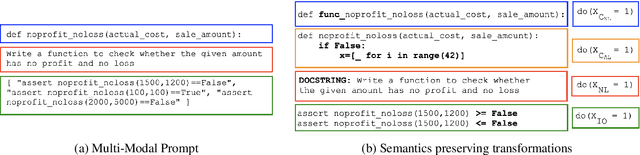
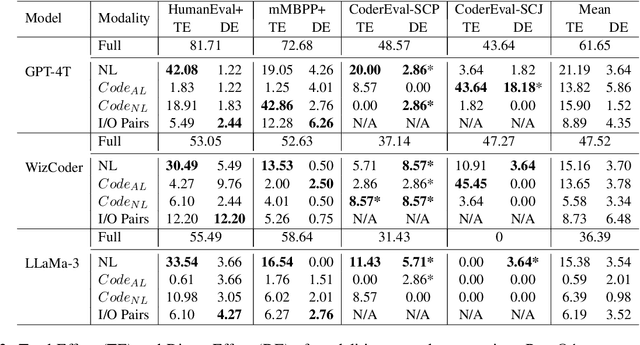
Abstract:In this paper, we propose CodeSCM, a Structural Causal Model (SCM) for analyzing multi-modal code generation using large language models (LLMs). By applying interventions to CodeSCM, we measure the causal effects of different prompt modalities, such as natural language, code, and input-output examples, on the model. CodeSCM introduces latent mediator variables to separate the code and natural language semantics of a multi-modal code generation prompt. Using the principles of Causal Mediation Analysis on these mediators we quantify direct effects representing the model's spurious leanings. We find that, in addition to natural language instructions, input-output examples significantly influence code generation.
Intent Detection and Entity Extraction from BioMedical Literature
Apr 04, 2024

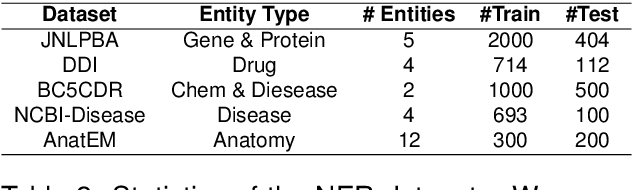
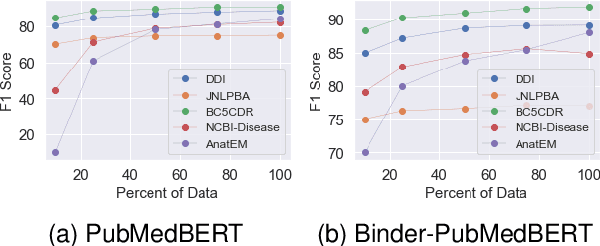
Abstract:Biomedical queries have become increasingly prevalent in web searches, reflecting the growing interest in accessing biomedical literature. Despite recent research on large-language models (LLMs) motivated by endeavours to attain generalized intelligence, their efficacy in replacing task and domain-specific natural language understanding approaches remains questionable. In this paper, we address this question by conducting a comprehensive empirical evaluation of intent detection and named entity recognition (NER) tasks from biomedical text. We show that Supervised Fine Tuned approaches are still relevant and more effective than general-purpose LLMs. Biomedical transformer models such as PubMedBERT can surpass ChatGPT on NER task with only 5 supervised examples.
TSO: Curriculum Generation using continuous optimization
Jun 16, 2021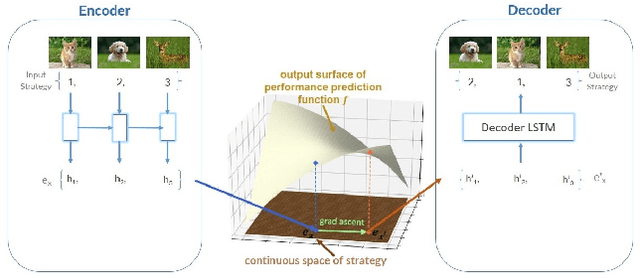
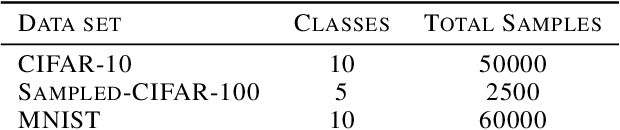


Abstract:The training of deep learning models poses vast challenges of including parameter tuning and ordering of training data. Significant research has been done in Curriculum learning for optimizing the sequence of training data. Recent works have focused on using complex reinforcement learning techniques to find the optimal data ordering strategy to maximize learning for a given network. In this paper, we present a simple and efficient technique based on continuous optimization. We call this new approach Training Sequence Optimization (TSO). There are three critical components in our proposed approach: (a) An encoder network maps/embeds training sequence into continuous space. (b) A predictor network uses the continuous representation of a strategy as input and predicts the accuracy for fixed network architecture. (c) A decoder further maps a continuous representation of a strategy to the ordered training dataset. The performance predictor and encoder enable us to perform gradient-based optimization in the continuous space to find the embedding of optimal training data ordering with potentially better accuracy. Experiments show that we can gain 2AP with our generated optimal curriculum strategy over the random strategy using the CIFAR-100 dataset and have better boosts than the state of the art CL algorithms. We do an ablation study varying the architecture, dataset and sample sizes showcasing our approach's robustness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge