Intent Detection and Entity Extraction from BioMedical Literature
Paper and Code
Apr 04, 2024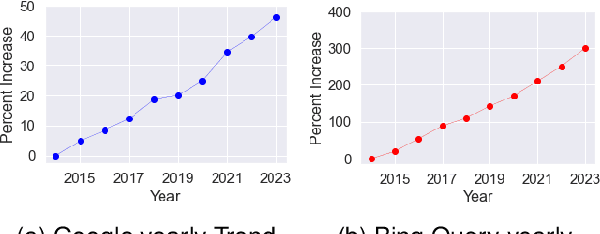

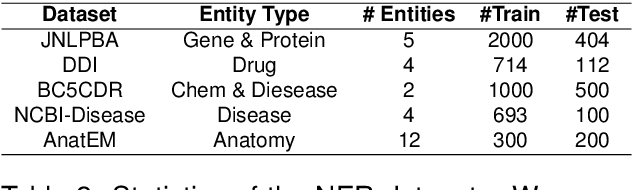
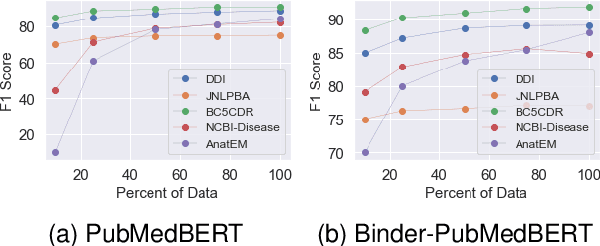
Biomedical queries have become increasingly prevalent in web searches, reflecting the growing interest in accessing biomedical literature. Despite recent research on large-language models (LLMs) motivated by endeavours to attain generalized intelligence, their efficacy in replacing task and domain-specific natural language understanding approaches remains questionable. In this paper, we address this question by conducting a comprehensive empirical evaluation of intent detection and named entity recognition (NER) tasks from biomedical text. We show that Supervised Fine Tuned approaches are still relevant and more effective than general-purpose LLMs. Biomedical transformer models such as PubMedBERT can surpass ChatGPT on NER task with only 5 supervised examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge