Mojtaba S. Fazli
Object Tracking in a $360^o$ View: A Novel Perspective on Bridging the Gap to Biomedical Advancements
Dec 02, 2024
Abstract:Object tracking is a fundamental tool in modern innovation, with applications in defense systems, autonomous vehicles, and biomedical research. It enables precise identification, monitoring, and spatiotemporal analysis of objects across sequential frames, providing insights into dynamic behaviors. In cell biology, object tracking is vital for uncovering cellular mechanisms, such as migration, interactions, and responses to drugs or pathogens. These insights drive breakthroughs in understanding disease progression and therapeutic interventions. Over time, object tracking methods have evolved from traditional feature-based approaches to advanced machine learning and deep learning frameworks. While classical methods are reliable in controlled settings, they struggle in complex environments with occlusions, variable lighting, and high object density. Deep learning models address these challenges by delivering greater accuracy, adaptability, and robustness. This review categorizes object tracking techniques into traditional, statistical, feature-based, and machine learning paradigms, with a focus on biomedical applications. These methods are essential for tracking cells and subcellular structures, advancing our understanding of health and disease. Key performance metrics, including accuracy, efficiency, and adaptability, are discussed. The paper explores limitations of current methods and highlights emerging trends to guide the development of next-generation tracking systems for biomedical research and broader scientific domains.
Artifact-Tolerant Clustering-Guided Contrastive Embedding Learning for Ophthalmic Images
Sep 02, 2022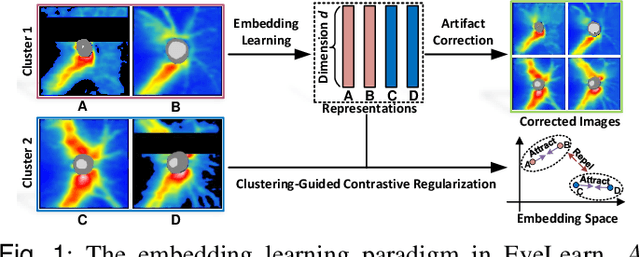
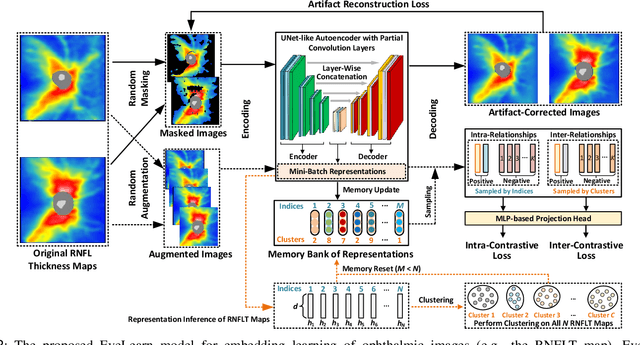
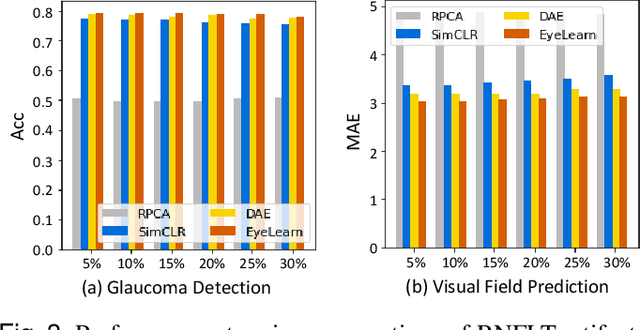
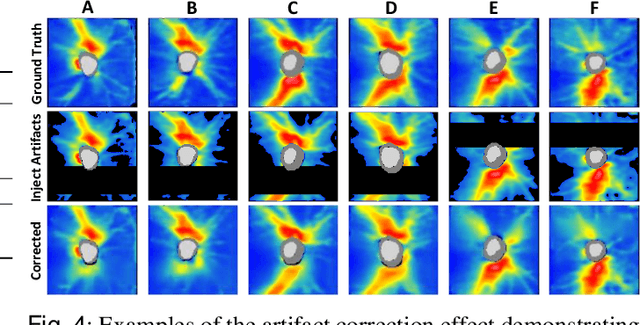
Abstract:Ophthalmic images and derivatives such as the retinal nerve fiber layer (RNFL) thickness map are crucial for detecting and monitoring ophthalmic diseases (e.g., glaucoma). For computer-aided diagnosis of eye diseases, the key technique is to automatically extract meaningful features from ophthalmic images that can reveal the biomarkers (e.g., RNFL thinning patterns) linked to functional vision loss. However, representation learning from ophthalmic images that links structural retinal damage with human vision loss is non-trivial mostly due to large anatomical variations between patients. The task becomes even more challenging in the presence of image artifacts, which are common due to issues with image acquisition and automated segmentation. In this paper, we propose an artifact-tolerant unsupervised learning framework termed EyeLearn for learning representations of ophthalmic images. EyeLearn has an artifact correction module to learn representations that can best predict artifact-free ophthalmic images. In addition, EyeLearn adopts a clustering-guided contrastive learning strategy to explicitly capture the intra- and inter-image affinities. During training, images are dynamically organized in clusters to form contrastive samples in which images in the same or different clusters are encouraged to learn similar or dissimilar representations, respectively. To evaluate EyeLearn, we use the learned representations for visual field prediction and glaucoma detection using a real-world ophthalmic image dataset of glaucoma patients. Extensive experiments and comparisons with state-of-the-art methods verified the effectiveness of EyeLearn for learning optimal feature representations from ophthalmic images.
Lightweight and Scalable Particle Tracking and Motion Clustering of 3D Cell Trajectories
Aug 14, 2019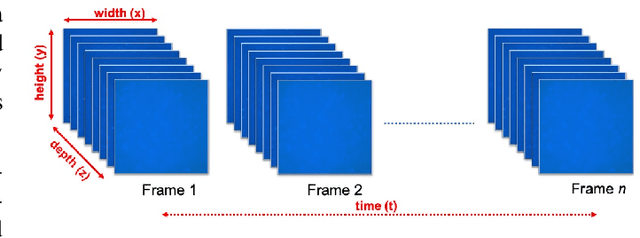
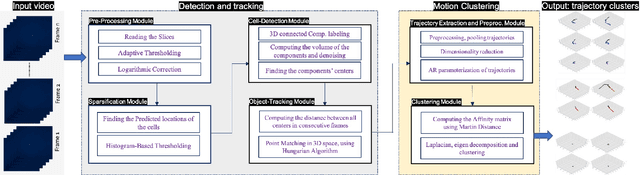
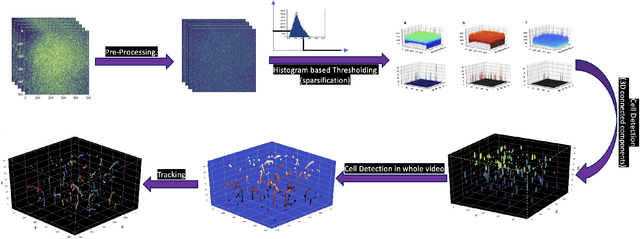
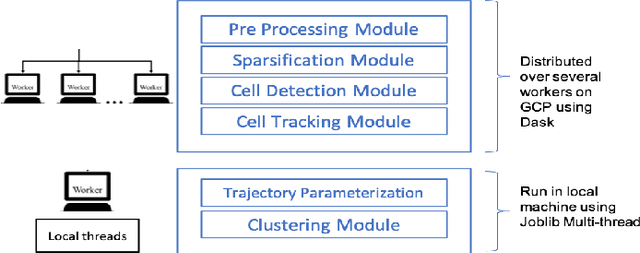
Abstract:Tracking cell particles in 3D microscopy videos is a challenging task but is of great significance for modeling the motion of cells. Proper characterization of the cell's shape, evolution, and their movement over time is crucial to understanding and modeling the mechanobiology of cell migration in many diseases. One in particular, toxoplasmosis is the disease caused by the parasite Toxoplasma gondii. Roughly, one-third of the world's population tests positive for T. gondii. Its virulence is linked to its lytic cycle, predicated on its motility and ability to enter and exit nucleated cells; therefore, studies elucidating its motility patterns are critical to the eventual development of therapeutic strategies. Here, we present a computational framework for fast and scalable detection, tracking, and identification of T. gondii motion phenotypes in 3D videos, in a completely unsupervised fashion. Our pipeline consists of several different modules including preprocessing, sparsification, cell detection, cell tracking, trajectories extraction, parametrization of the trajectories; and finally, a clustering step. Additionally, we identified the computational bottlenecks, and developed a lightweight and highly scalable pipeline through a combination of task distribution and parallelism. Our results prove both the accuracy and performance of our method.
Unsupervised Discovery of Toxoplasma gondii Motility Phenotypes
Jan 11, 2018



Abstract:Toxoplasma gondii is a parasitic protozoan that causes dis- seminated toxoplasmosis, a disease that afflicts roughly a third of the worlds population. Its virulence is predicated on its motility and ability to enter and exit nucleated cells; therefore, studies elucidating its mechanism of motility and in particular, its motility patterns in the context of its lytic cycle, are critical to the eventual development of therapeutic strate- gies. Here, we present an end-to-end computational pipeline for identifying T. gondii motility phenotypes in a completely unsupervised, data-driven way. We track the parasites before and after addition of extracellular Ca2+ to study its effects on the parasite motility patterns and use this information to parameterize the motion and group it according to similarity of spatiotemporal dynamics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge