Shannon Quinn
Object Tracking in a $360^o$ View: A Novel Perspective on Bridging the Gap to Biomedical Advancements
Dec 02, 2024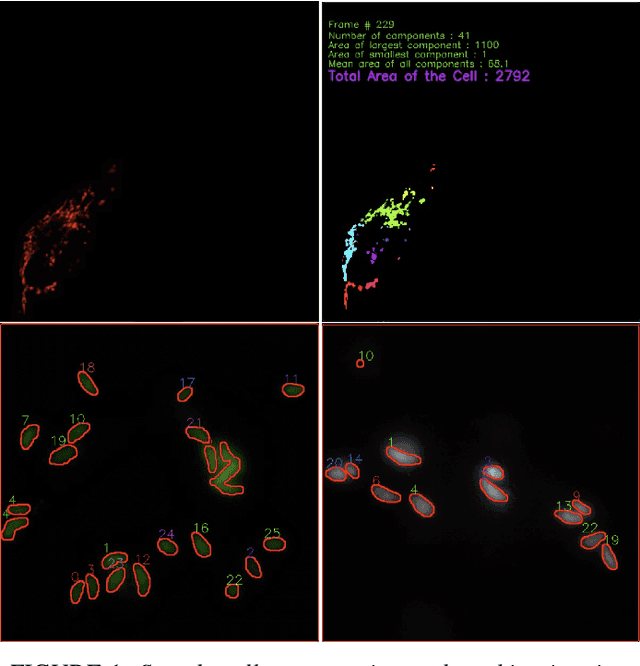
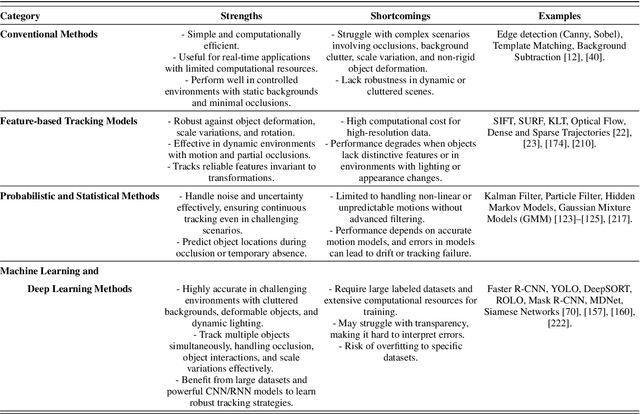
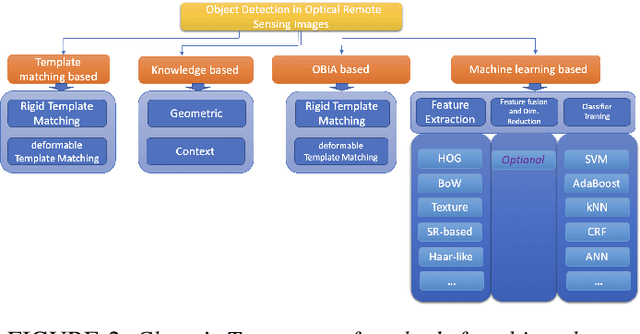
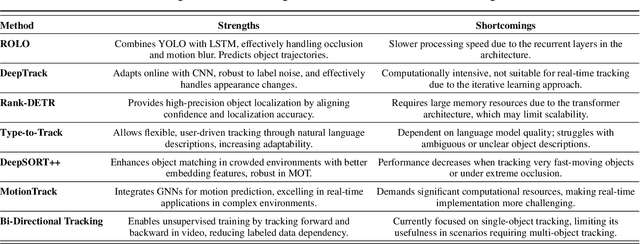
Abstract:Object tracking is a fundamental tool in modern innovation, with applications in defense systems, autonomous vehicles, and biomedical research. It enables precise identification, monitoring, and spatiotemporal analysis of objects across sequential frames, providing insights into dynamic behaviors. In cell biology, object tracking is vital for uncovering cellular mechanisms, such as migration, interactions, and responses to drugs or pathogens. These insights drive breakthroughs in understanding disease progression and therapeutic interventions. Over time, object tracking methods have evolved from traditional feature-based approaches to advanced machine learning and deep learning frameworks. While classical methods are reliable in controlled settings, they struggle in complex environments with occlusions, variable lighting, and high object density. Deep learning models address these challenges by delivering greater accuracy, adaptability, and robustness. This review categorizes object tracking techniques into traditional, statistical, feature-based, and machine learning paradigms, with a focus on biomedical applications. These methods are essential for tracking cells and subcellular structures, advancing our understanding of health and disease. Key performance metrics, including accuracy, efficiency, and adaptability, are discussed. The paper explores limitations of current methods and highlights emerging trends to guide the development of next-generation tracking systems for biomedical research and broader scientific domains.
Lightweight and Scalable Particle Tracking and Motion Clustering of 3D Cell Trajectories
Aug 14, 2019
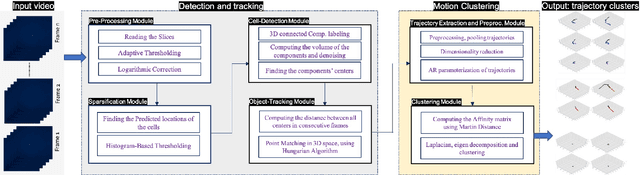
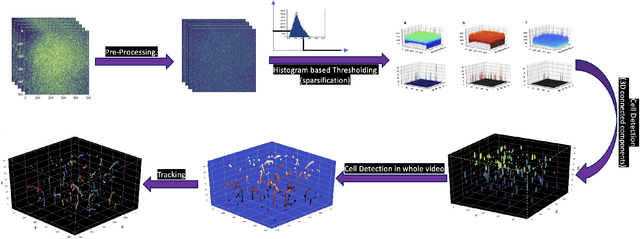
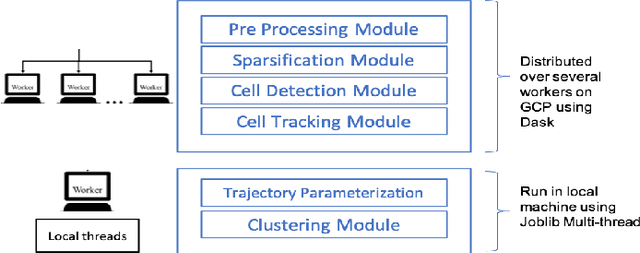
Abstract:Tracking cell particles in 3D microscopy videos is a challenging task but is of great significance for modeling the motion of cells. Proper characterization of the cell's shape, evolution, and their movement over time is crucial to understanding and modeling the mechanobiology of cell migration in many diseases. One in particular, toxoplasmosis is the disease caused by the parasite Toxoplasma gondii. Roughly, one-third of the world's population tests positive for T. gondii. Its virulence is linked to its lytic cycle, predicated on its motility and ability to enter and exit nucleated cells; therefore, studies elucidating its motility patterns are critical to the eventual development of therapeutic strategies. Here, we present a computational framework for fast and scalable detection, tracking, and identification of T. gondii motion phenotypes in 3D videos, in a completely unsupervised fashion. Our pipeline consists of several different modules including preprocessing, sparsification, cell detection, cell tracking, trajectories extraction, parametrization of the trajectories; and finally, a clustering step. Additionally, we identified the computational bottlenecks, and developed a lightweight and highly scalable pipeline through a combination of task distribution and parallelism. Our results prove both the accuracy and performance of our method.
Towards a Decentralized, Autonomous Multiagent Framework for Mitigating Crop Loss
Jan 07, 2019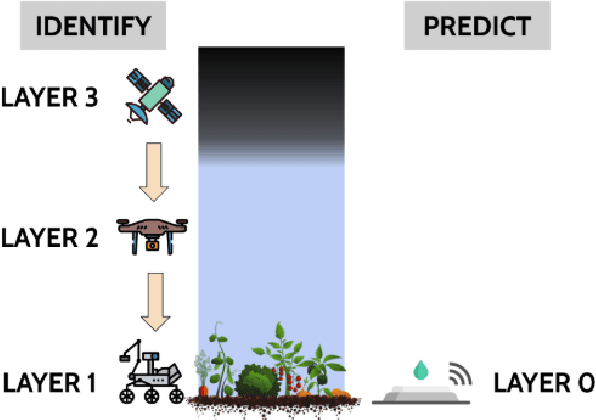
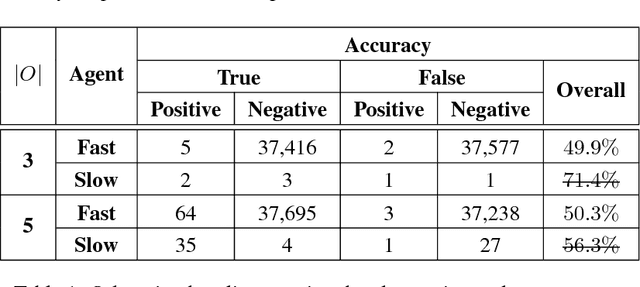
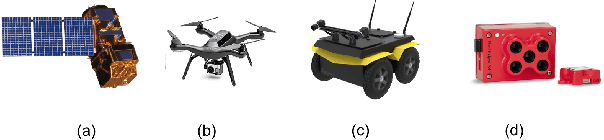
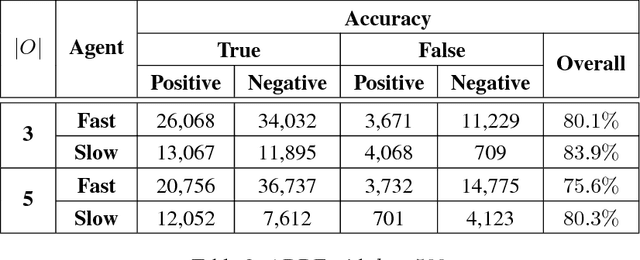
Abstract:We propose a generalized decision-theoretic system for a heterogeneous team of autonomous agents who are tasked with online identification of phenotypically expressed stress in crop fields.. This system employs four distinct types of agents, specific to four available sensor modalities: satellites (Layer 3), uninhabited aerial vehicles (L2), uninhabited ground vehicles (L1), and static ground-level sensors (L0). Layers 3, 2, and 1 are tasked with performing image processing at the available resolution of the sensor modality and, along with data generated by layer 0 sensors, identify erroneous differences that arise over time. Our goal is to limit the use of the more computationally and temporally expensive subsequent layers. Therefore, from layer 3 to 1, each layer only investigates areas that previous layers have identified as potentially afflicted by stress. We introduce a reinforcement learning technique based on Perkins' Monte Carlo Exploring Starts for a generalized Markovian model for each layer's decision problem, and label the system the Agricultural Distributed Decision Framework (ADDF). As our domain is real-world and online, we illustrate implementations of the two major components of our system: a clustering-based image processing methodology and a two-layer POMDP implementation.
Generative Spatiotemporal Modeling Of Neutrophil Behavior
Apr 02, 2018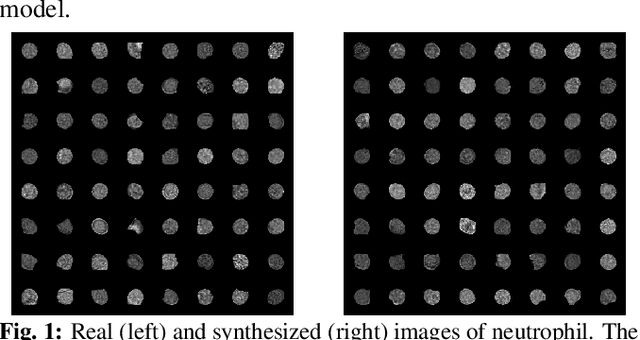
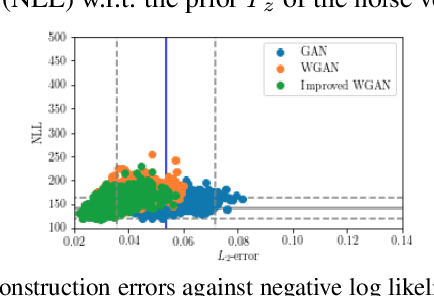

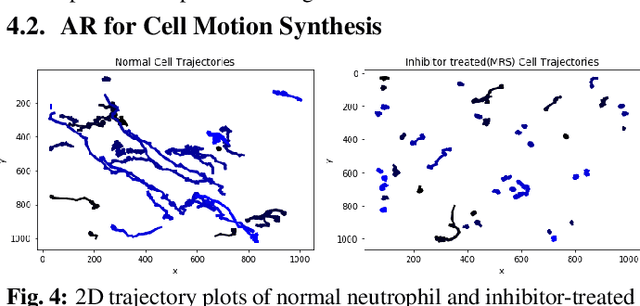
Abstract:Cell motion and appearance have a strong correlation with cell cycle and disease progression. Many contemporary efforts in machine learning utilize spatio-temporal models to predict a cell's physical state and, consequently, the advancement of disease. Alternatively, generative models learn the underlying distribution of the data, creating holistic representations that can be used in learning. In this work, we propose an aggregate model that combine Generative Adversarial Networks (GANs) and Autoregressive (AR) models to predict cell motion and appearance in human neutrophils imaged by differential interference contrast (DIC) microscopy. We bifurcate the task of learning cell statistics by leveraging GANs for the spatial component and AR models for the temporal component. The aggregate model learned results offer a promising computational environment for studying changes in organellar shape, quantity, and spatial distribution over large sequences.
Unsupervised Discovery of Toxoplasma gondii Motility Phenotypes
Jan 11, 2018



Abstract:Toxoplasma gondii is a parasitic protozoan that causes dis- seminated toxoplasmosis, a disease that afflicts roughly a third of the worlds population. Its virulence is predicated on its motility and ability to enter and exit nucleated cells; therefore, studies elucidating its mechanism of motility and in particular, its motility patterns in the context of its lytic cycle, are critical to the eventual development of therapeutic strate- gies. Here, we present an end-to-end computational pipeline for identifying T. gondii motility phenotypes in a completely unsupervised, data-driven way. We track the parasites before and after addition of extracellular Ca2+ to study its effects on the parasite motility patterns and use this information to parameterize the motion and group it according to similarity of spatiotemporal dynamics.
Computational Motility Tracking of Calcium Dynamics in Toxoplasma gondii
Aug 18, 2017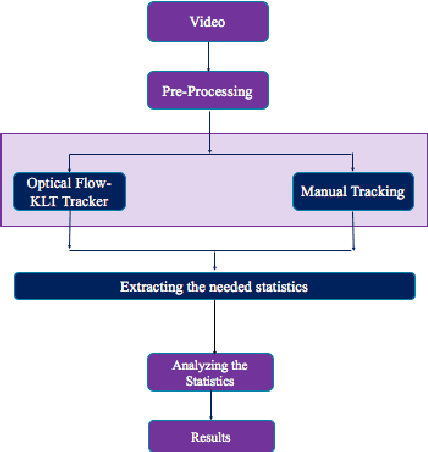
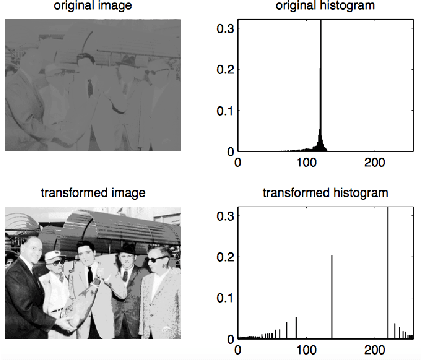
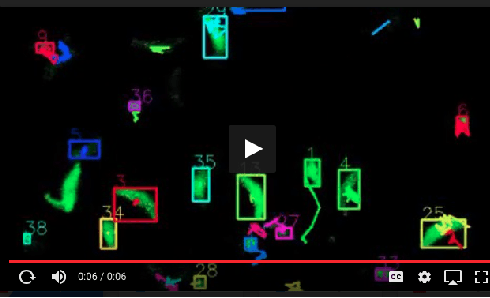
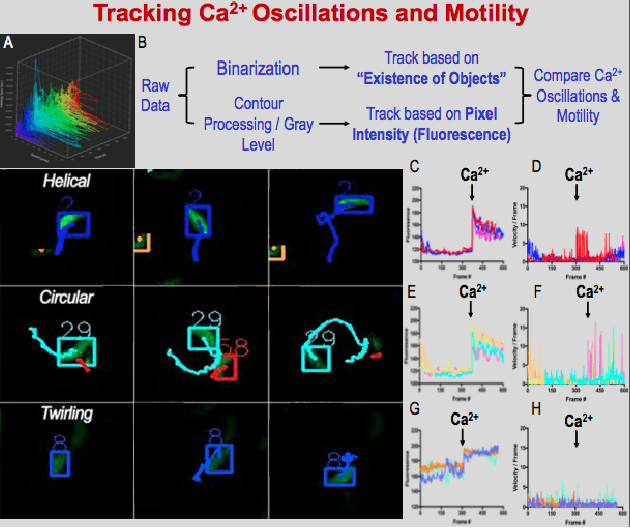
Abstract:Toxoplasma gondii is the causative agent responsible for toxoplasmosis and serves as one of the most common parasites in the world. For a successful lytic cycle, T. gondii must traverse biological barriers in order to invade host cells, and as such, motility is critical for its virulence. Calcium signaling, governed by fluctuations in cytosolic calcium (Ca2+) concentrations, is utilized universally across life and regulates many cellular processes, including the stimulation of T. gondii virulence factors such as motility. Therefore, increases in cytosolic calcium, called calcium oscillations, serve as a means to link and quantify the intracellular signaling processes that lead to T. gondii motility and invasion. Here, we describe our work extracting, quantifying and modeling motility patterns of T. gondii before and after the addition of pharmacological drugs and/or extracellular calcium. We demonstrate a computational pipeline including a robust tracking system using optical flow and dense trajectory features to extract T. gondii motility patterns. Using this pipeline, we were able to track changes in T.gondii motility in response to cytosolic Ca2+ fluxes in extracellular parasites. This allows us to study how Ca2+ signaling via release from intracellular Ca2+ stores and/or from extracellular Ca2+ entry relates to motility patterns, a crucial first step in developing countermeasures for T. gondii virulence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge