Narita Pandhe
Generative Spatiotemporal Modeling Of Neutrophil Behavior
Apr 02, 2018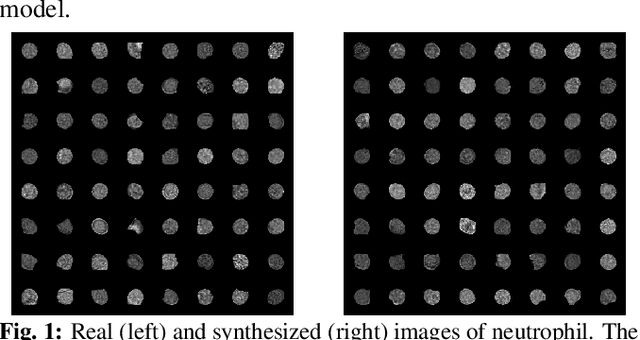
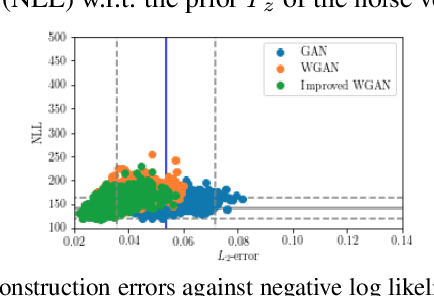

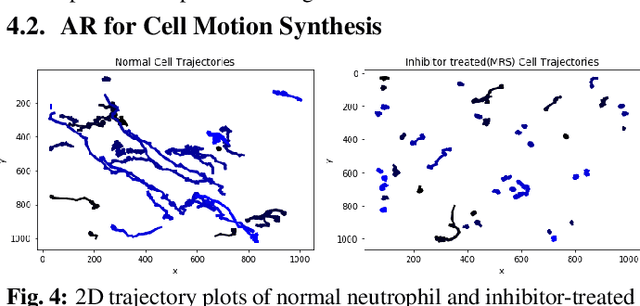
Abstract:Cell motion and appearance have a strong correlation with cell cycle and disease progression. Many contemporary efforts in machine learning utilize spatio-temporal models to predict a cell's physical state and, consequently, the advancement of disease. Alternatively, generative models learn the underlying distribution of the data, creating holistic representations that can be used in learning. In this work, we propose an aggregate model that combine Generative Adversarial Networks (GANs) and Autoregressive (AR) models to predict cell motion and appearance in human neutrophils imaged by differential interference contrast (DIC) microscopy. We bifurcate the task of learning cell statistics by leveraging GANs for the spatial component and AR models for the temporal component. The aggregate model learned results offer a promising computational environment for studying changes in organellar shape, quantity, and spatial distribution over large sequences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge