Mikkel Fly Kragh
Development and validation of deep learning based embryo selection across multiple days of transfer
Oct 05, 2022



Abstract:This work describes the development and validation of a fully automated deep learning model, iDAScore v2.0, for the evaluation of embryos incubated for 2, 3, and 5 or more days. The model is trained and evaluated on an extensive and diverse dataset including 181,428 embryos from 22 IVF clinics across the world. For discriminating transferred embryos with known outcome (KID), we show AUCs ranging from 0.621 to 0.708 depending on the day of transfer. Predictive performance increased over time and showed a strong correlation with morphokinetic parameters. The model has equivalent performance to KIDScore D3 on day 3 embryos while significantly surpassing the performance of KIDScore D5 v3 on day 5+ embryos. This model provides an analysis of time-lapse sequences without the need for user input, and provides a reliable method for ranking embryos for likelihood to implant, at both cleavage and blastocyst stages. This greatly improves embryo grading consistency and saves time compared to traditional embryo evaluation methods.
Robust and generalizable embryo selection based on artificial intelligence and time-lapse image sequences
Mar 12, 2021



Abstract:Assessing and selecting the most viable embryos for transfer is an essential part of in vitro fertilization (IVF). In recent years, several approaches have been made to improve and automate the procedure using artificial intelligence (AI) and deep learning. Based on images of embryos with known implantation data (KID), AI models have been trained to automatically score embryos related to their chance of achieving a successful implantation. However, as of now, only limited research has been conducted to evaluate how embryo selection models generalize to new clinics and how they perform in subgroup analyses across various conditions. In this paper, we investigate how a deep learning-based embryo selection model using only time-lapse image sequences performs across different patient ages and clinical conditions, and how it correlates with traditional morphokinetic parameters. The model was trained and evaluated based on a large dataset from 18 IVF centers consisting of 115,832 embryos, of which 14,644 embryos were transferred KID embryos. In an independent test set, the AI model sorted KID embryos with an area under the curve (AUC) of a receiver operating characteristic curve of 0.67 and all embryos with an AUC of 0.95. A clinic hold-out test showed that the model generalized to new clinics with an AUC range of 0.60-0.75 for KID embryos. Across different subgroups of age, insemination method, incubation time, and transfer protocol, the AUC ranged between 0.63 and 0.69. Furthermore, model predictions correlated positively with blastocyst grading and negatively with direct cleavages. The fully automated iDAScore v1.0 model was shown to perform at least as good as a state-of-the-art manual embryo selection model. Moreover, full automatization of embryo scoring implies fewer manual evaluations and eliminates biases due to inter- and intraobserver variation.
UnsuperPoint: End-to-end Unsupervised Interest Point Detector and Descriptor
Jul 09, 2019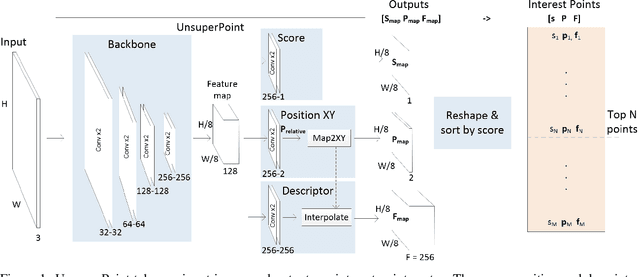
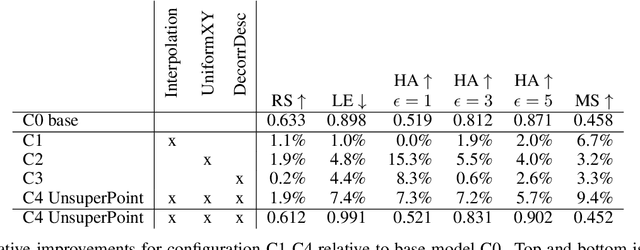
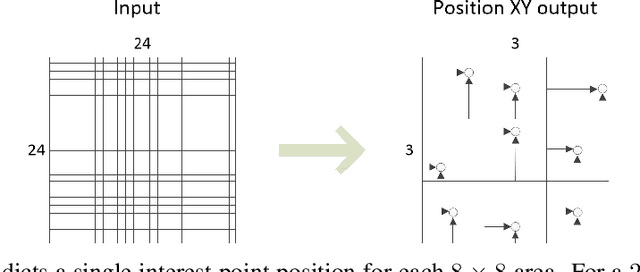
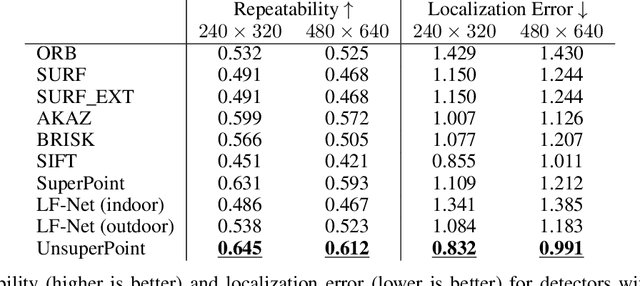
Abstract:It is hard to create consistent ground truth data for interest points in natural images, since interest points are hard to define clearly and consistently for a human annotator. This makes interest point detectors non-trivial to build. In this work, we introduce an unsupervised deep learning-based interest point detector and descriptor. Using a self-supervised approach, we utilize a siamese network and a novel loss function that enables interest point scores and positions to be learned automatically. The resulting interest point detector and descriptor is UnsuperPoint. We use regression of point positions to 1) make UnsuperPoint end-to-end trainable and 2) to incorporate non-maximum suppression in the model. Unlike most trainable detectors, it requires no generation of pseudo ground truth points, no structure-from-motion-generated representations and the model is learned from only one round of training. Furthermore, we introduce a novel loss function to regularize network predictions to be uniformly distributed. UnsuperPoint runs in real-time with 323 frames per second (fps) at a resolution of $224\times320$ and 90 fps at $480\times640$. It is comparable or better than state-of-the-art performance when measured for speed, repeatability, localization, matching score and homography estimation on the HPatch dataset.
FieldSAFE: Dataset for Obstacle Detection in Agriculture
Sep 11, 2017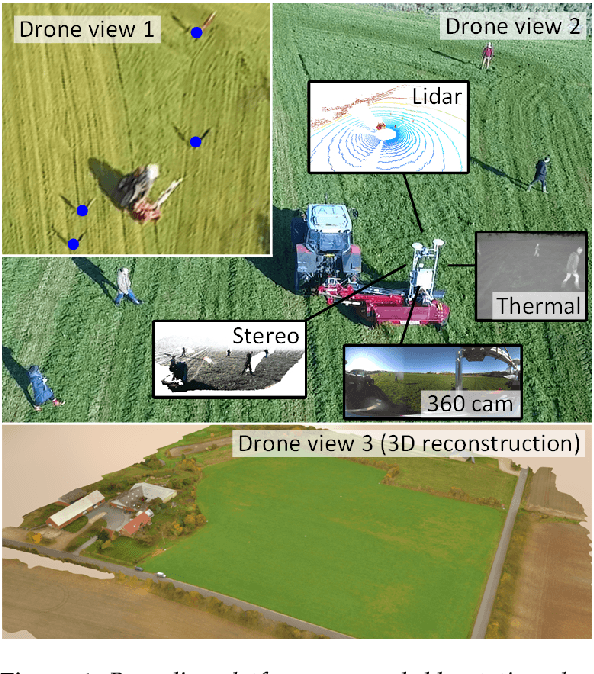

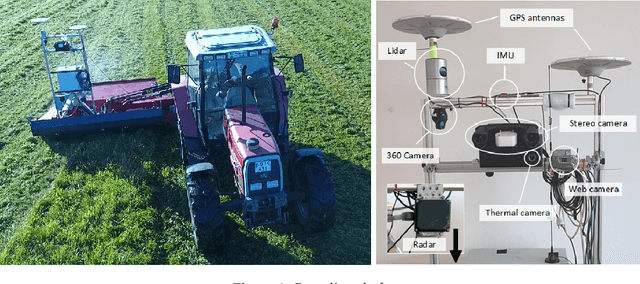

Abstract:In this paper, we present a novel multi-modal dataset for obstacle detection in agriculture. The dataset comprises approximately 2 hours of raw sensor data from a tractor-mounted sensor system in a grass mowing scenario in Denmark, October 2016. Sensing modalities include stereo camera, thermal camera, web camera, 360-degree camera, lidar, and radar, while precise localization is available from fused IMU and GNSS. Both static and moving obstacles are present including humans, mannequin dolls, rocks, barrels, buildings, vehicles, and vegetation. All obstacles have ground truth object labels and geographic coordinates.
* Submitted to special issue of MDPI Sensors: Sensors in Agriculture
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge