Mijung Kim
Hallucination by Code Generation LLMs: Taxonomy, Benchmarks, Mitigation, and Challenges
Apr 29, 2025



Abstract:Recent technical breakthroughs in large language models (LLMs) have enabled them to fluently generate source code. Software developers often leverage both general-purpose and code-specialized LLMs to revise existing code or even generate a whole function from scratch. These capabilities are also beneficial in no-code or low-code contexts, in which one can write programs without a technical background. However, due to their internal design, LLMs are prone to generating hallucinations, which are incorrect, nonsensical, and not justifiable information but difficult to identify its presence. This problem also occurs when generating source code. Once hallucinated code is produced, it is often challenging for users to identify and fix it, especially when such hallucinations can be identified under specific execution paths. As a result, the hallucinated code may remain unnoticed within the codebase. This survey investigates recent studies and techniques relevant to hallucinations generated by CodeLLMs. We categorize the types of hallucinations in the code generated by CodeLLMs, review existing benchmarks and mitigation strategies, and identify open challenges. Based on these findings, this survey outlines further research directions in the detection and removal of hallucinations produced by CodeLLMs.
Age Prediction From Face Images Via Contrastive Learning
Aug 23, 2023



Abstract:This paper presents a novel approach for accurately estimating age from face images, which overcomes the challenge of collecting a large dataset of individuals with the same identity at different ages. Instead, we leverage readily available face datasets of different people at different ages and aim to extract age-related features using contrastive learning. Our method emphasizes these relevant features while suppressing identity-related features using a combination of cosine similarity and triplet margin losses. We demonstrate the effectiveness of our proposed approach by achieving state-of-the-art performance on two public datasets, FG-NET and MORPH-II.
Web Applicable Computer-aided Diagnosis of Glaucoma Using Deep Learning
Dec 06, 2018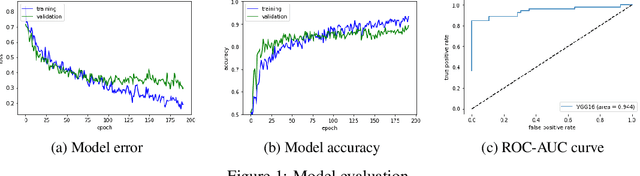

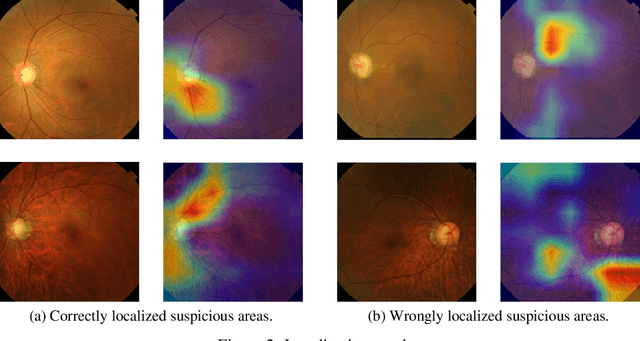
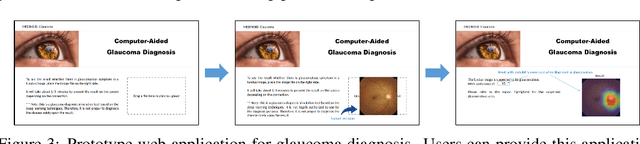
Abstract:Glaucoma is a major eye disease, leading to vision loss in the absence of proper medical treatment. Current diagnosis of glaucoma is performed by ophthalmologists who are often analyzing several types of medical images generated by different types of medical equipment. Capturing and analyzing these medical images is labor-intensive and expensive. In this paper, we present a novel computational approach towards glaucoma diagnosis and localization, only making use of eye fundus images that are analyzed by state-of-the-art deep learning techniques. Specifically, our approach leverages Convolutional Neural Networks (CNNs) and Gradient-weighted Class Activation Mapping (Grad-CAM) for glaucoma diagnosis and localization, respectively. Quantitative and qualitative results, as obtained for a small-sized dataset with no segmentation ground truth, demonstrate that the proposed approach is promising, for instance achieving an accuracy of 0.91$\pm0.02$ and an ROC-AUC score of 0.94 for the diagnosis task. Furthermore, we present a publicly available prototype web application that integrates our predictive model, with the goal of making effective glaucoma diagnosis available to a wide audience.
Interpretable Convolutional Neural Networks for Effective Translation Initiation Site Prediction
Nov 27, 2017

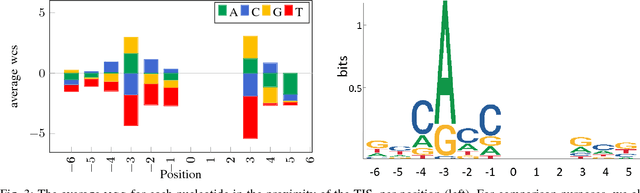
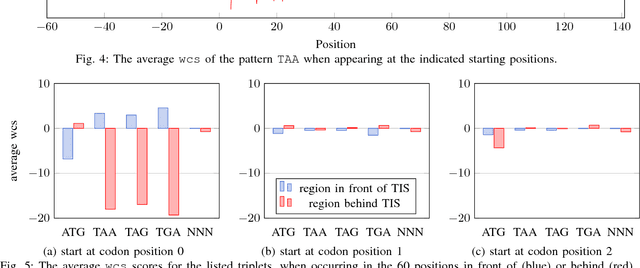
Abstract:Thanks to rapidly evolving sequencing techniques, the amount of genomic data at our disposal is growing increasingly large. Determining the gene structure is a fundamental requirement to effectively interpret gene function and regulation. An important part in that determination process is the identification of translation initiation sites. In this paper, we propose a novel approach for automatic prediction of translation initiation sites, leveraging convolutional neural networks that allow for automatic feature extraction. Our experimental results demonstrate that we are able to improve the state-of-the-art approaches with a decrease of 75.2% in false positive rate and with a decrease of 24.5% in error rate on chosen datasets. Furthermore, an in-depth analysis of the decision-making process used by our predictive model shows that our neural network implicitly learns biologically relevant features from scratch, without any prior knowledge about the problem at hand, such as the Kozak consensus sequence, the influence of stop and start codons in the sequence and the presence of donor splice site patterns. In summary, our findings yield a better understanding of the internal reasoning of a convolutional neural network when applying such a neural network to genomic data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge