Miguel Romero
The Distributional Uncertainty of the SHAP score in Explainable Machine Learning
Jan 23, 2024



Abstract:Attribution scores reflect how important the feature values in an input entity are for the output of a machine learning model. One of the most popular attribution scores is the SHAP score, which is an instantiation of the general Shapley value used in coalition game theory. The definition of this score relies on a probability distribution on the entity population. Since the exact distribution is generally unknown, it needs to be assigned subjectively or be estimated from data, which may lead to misleading feature scores. In this paper, we propose a principled framework for reasoning on SHAP scores under unknown entity population distributions. In our framework, we consider an uncertainty region that contains the potential distributions, and the SHAP score of a feature becomes a function defined over this region. We study the basic problems of finding maxima and minima of this function, which allows us to determine tight ranges for the SHAP scores of all features. In particular, we pinpoint the complexity of these problems, and other related ones, showing them to be NP-complete. Finally, we present experiments on a real-world dataset, showing that our framework may contribute to a more robust feature scoring.
A neuro-symbolic framework for answering conjunctive queries
Oct 06, 2023Abstract:The problem of answering logical queries over incomplete knowledge graphs is receiving significant attention in the machine learning community. Neuro-symbolic models are a promising recent approach, showing good performance and allowing for good interpretability properties. These models rely on trained architectures to execute atomic queries, combining them with modules that simulate the symbolic operators in queries. Unfortunately, most neuro-symbolic query processors are limited to the so-called tree-like logical queries that admit a bottom-up execution, where the leaves are constant values or anchors, and the root is the target variable. Tree-like queries, while expressive, fail short to express properties in knowledge graphs that are important in practice, such as the existence of multiple edges between entities or the presence of triangles. We propose a framework for answering arbitrary conjunctive queries over incomplete knowledge graphs. The main idea of our method is to approximate a cyclic query by an infinite family of tree-like queries, and then leverage existing models for the latter. Our approximations achieve strong guarantees: they are complete, i.e. there are no false negatives, and optimal, i.e. they provide the best possible approximation using tree-like queries. Our method requires the approximations to be tree-like queries where the leaves are anchors or existentially quantified variables. Hence, we also show how some of the existing neuro-symbolic models can handle these queries, which is of independent interest. Experiments show that our approximation strategy achieves competitive results, and that including queries with existentially quantified variables tends to improve the general performance of these models, both on tree-like queries and on our approximation strategy.
Hierarchy exploitation to detect missing annotations on hierarchical multi-label classification
Jul 13, 2022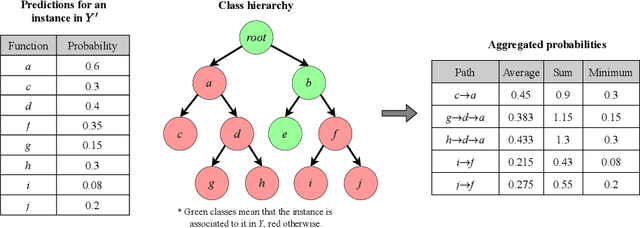
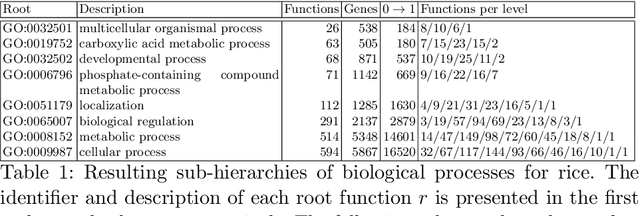
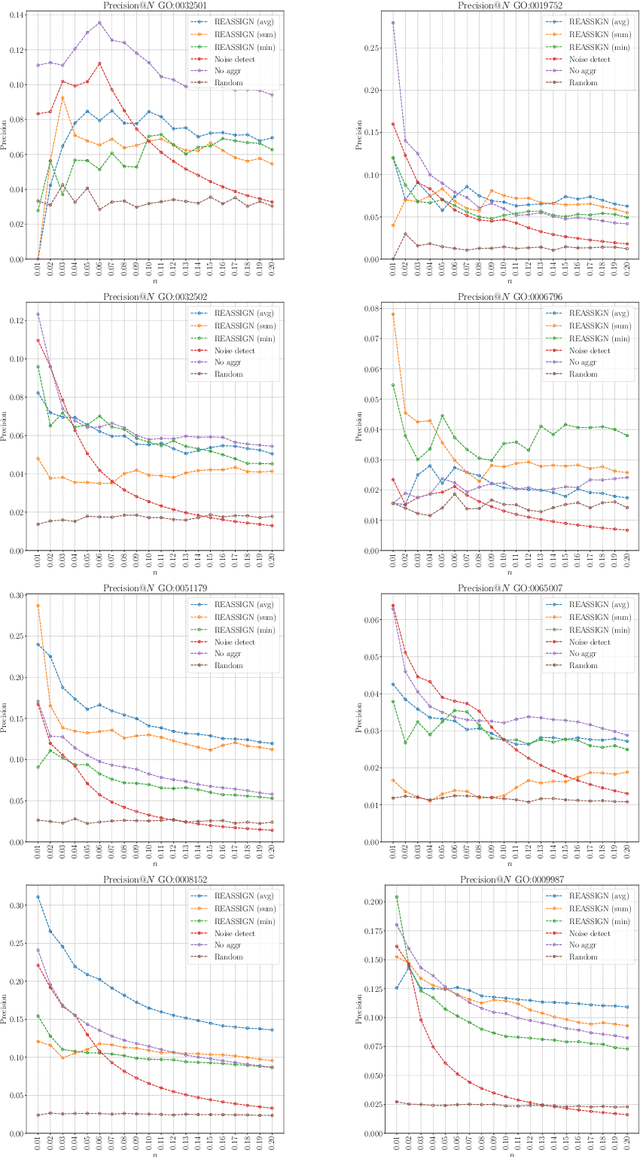
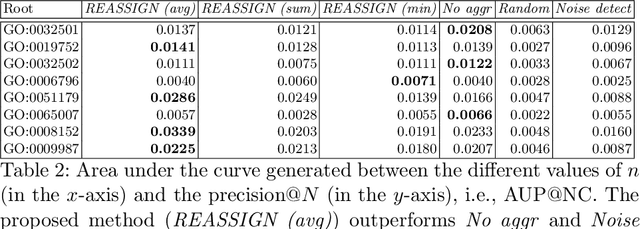
Abstract:The availability of genomic data has grown exponentially in the last decade, mainly due to the development of new sequencing technologies. Based on the interactions between genes (and gene products) extracted from the increasing genomic data, numerous studies have focused on the identification of associations between genes and functions. While these studies have shown great promise, the problem of annotating genes with functions remains an open challenge. In this work, we present a method to detect missing annotations in hierarchical multi-label classification datasets. We propose a method that exploits the class hierarchy by computing aggregated probabilities to the paths of classes from the leaves to the root for each instance. The proposed method is presented in the context of predicting missing gene function annotations, where these aggregated probabilities are further used to select a set of annotations to be verified through in vivo experiments. The experiments on Oriza sativa Japonica, a variety of rice, showcase that incorporating the hierarchy of classes into the method often improves the predictive performance and our proposed method yields superior results when compared to competitor methods from the literature.
Feature extraction using Spectral Clustering for Gene Function Prediction
Mar 25, 2022



Abstract:Gene annotation addresses the problem of predicting unknown associations between gene and functions (e.g., biological processes) of a specific organism. Despite recent advances, the cost and time demanded by annotation procedures that rely largely on in vivo biological experiments remain prohibitively high. This paper presents a novel in silico approach for to the annotation problem that combines cluster analysis and hierarchical multi-label classification (HMC). The approach uses spectral clustering to extract new features from the gene co-expression network (GCN) and enrich the prediction task. HMC is used to build multiple estimators that consider the hierarchical structure of gene functions. The proposed approach is applied to a case study on Zea mays, one of the most dominant and productive crops in the world. The results illustrate how in silico approaches are key to reduce the time and costs of gene annotation. More specifically, they highlight the importance of: (i) building new features that represent the structure of gene relationships in GCNs to annotate genes; and (ii) taking into account the structure of biological processes to obtain consistent predictions.
A Top-down Supervised Learning Approach to Hierarchical Multi-label Classification in Networks
Mar 23, 2022



Abstract:Node classification is the task of inferring or predicting missing node attributes from information available for other nodes in a network. This paper presents a general prediction model to hierarchical multi-label classification (HMC), where the attributes to be inferred can be specified as a strict poset. It is based on a top-down classification approach that addresses hierarchical multi-label classification with supervised learning by building a local classifier per class. The proposed model is showcased with a case study on the prediction of gene functions for Oryza sativa Japonica, a variety of rice. It is compared to the Hierarchical Binomial-Neighborhood, a probabilistic model, by evaluating both approaches in terms of prediction performance and computational cost. The results in this work support the working hypothesis that the proposed model can achieve good levels of prediction efficiency, while scaling up in relation to the state of the art.
Spectral Evolution with Approximated Eigenvalue Trajectories for Link Prediction
Jun 22, 2020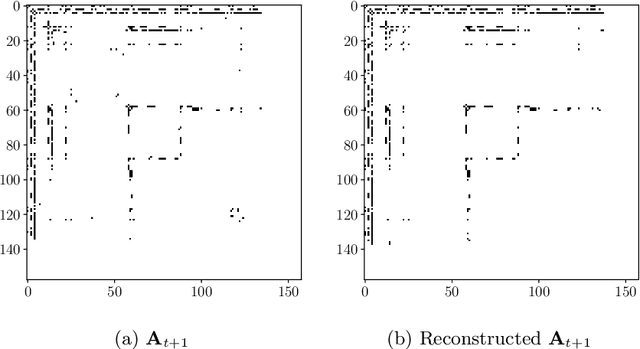



Abstract:The spectral evolution model aims to characterize the growth of large networks (i.e., how they evolve as new edges are established) in terms of the eigenvalue decomposition of the adjacency matrices. It assumes that, while eigenvectors remain constant, eigenvalues evolve in a predictable manner over time. This paper extends the original formulation of the model twofold. First, it presents a method to compute an approximation of the spectral evolution of eigenvalues based on the Rayleigh quotient. Second, it proposes an algorithm to estimate the evolution of eigenvalues by extrapolating only a fraction of their approximated values. The proposed model is used to characterize mention networks of users who posted tweets that include the most popular political hashtags in Colombia from August 2017 to August 2018 (the period which concludes the disarmament of the Revolutionary Armed Forces of Colombia). To evaluate the extent to which the spectral evolution model resembles these networks, link prediction methods based on learning algorithms (i.e., extrapolation and regression) and graph kernels are implemented. Experimental results show that the learning algorithms deployed on the approximated trajectories outperform the usual kernel and extrapolation methods at predicting the formation of new edges.
Training Deep Learning models with small datasets
Dec 14, 2019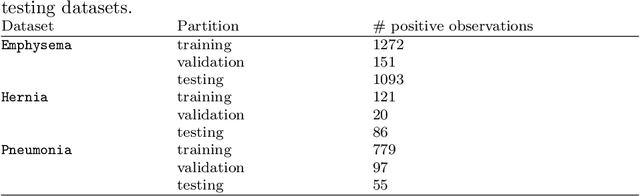
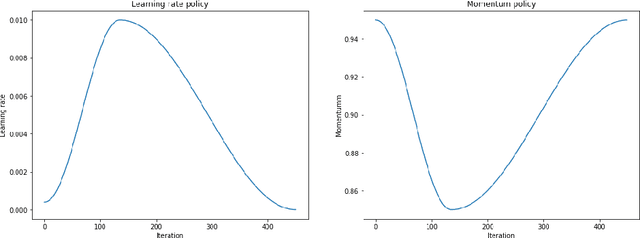
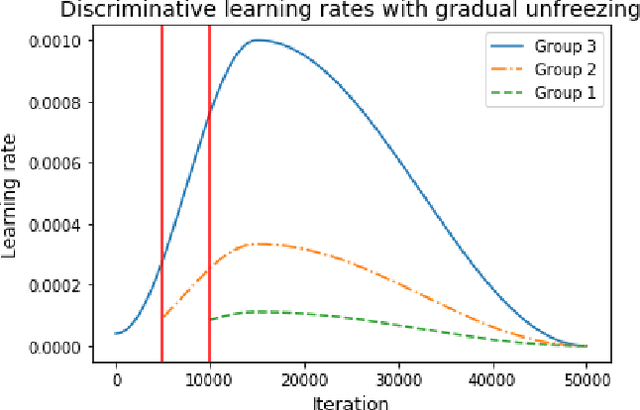
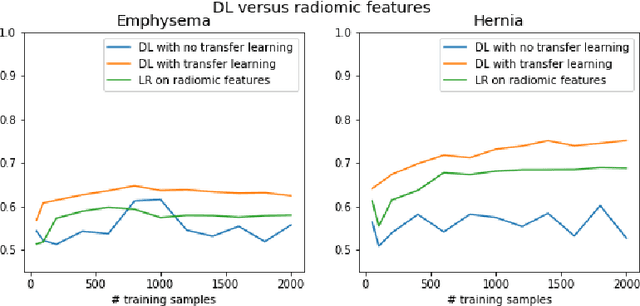
Abstract:The growing use of Machine Learning has produced significant advances in many fields. For image-based tasks, however, the use of deep learning remains challenging in small datasets. In this article, we review, evaluate and compare the current state of the art techniques in training neural networks to elucidate which techniques work best for small datasets. We further propose a path forward for the improvement of model accuracy in medical imaging applications. We observed best results from one cycle training, discriminative learning rates with gradual freezing and parameter modification after transfer learning. We also established that when datasets are small, transfer learning plays an important role beyond parameter initialization by reusing previously learned features. Surprisingly we observed that there is little advantage in using pre-trained networks in images from another part of the body compared to Imagenet. On the contrary, if images from the same part of the body are available then transfer learning can produce a significant improvement in performance with as little as 50 images in the training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge