Michael Xiao
PCS-UQ: Uncertainty Quantification via the Predictability-Computability-Stability Framework
May 13, 2025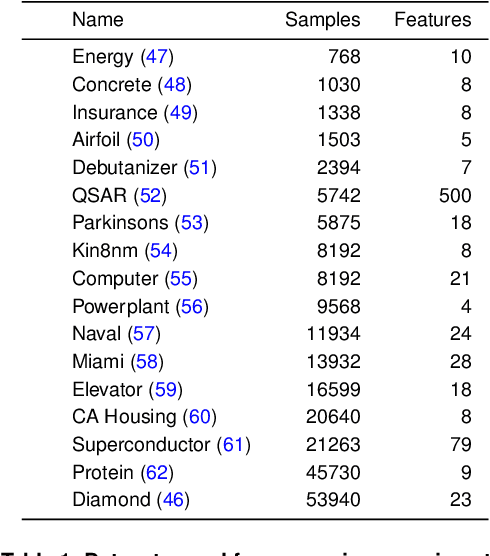
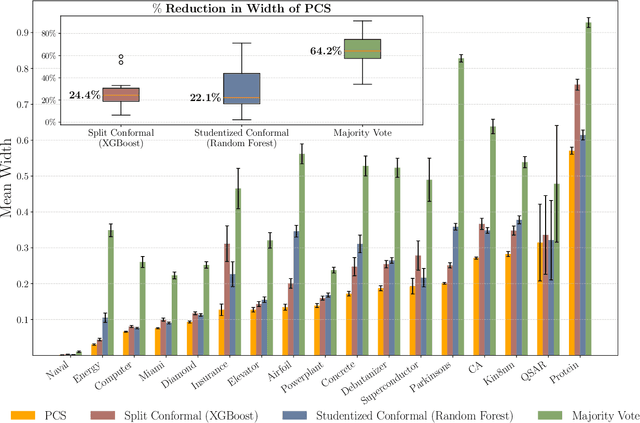
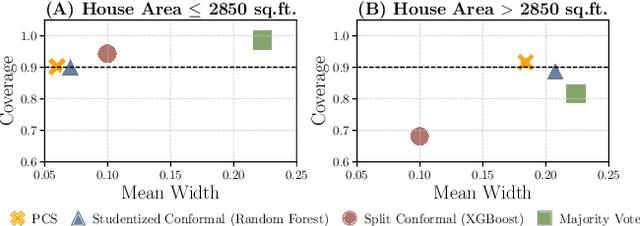
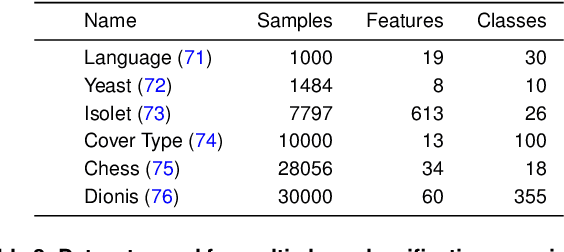
Abstract:As machine learning (ML) models are increasingly deployed in high-stakes domains, trustworthy uncertainty quantification (UQ) is critical for ensuring the safety and reliability of these models. Traditional UQ methods rely on specifying a true generative model and are not robust to misspecification. On the other hand, conformal inference allows for arbitrary ML models but does not consider model selection, which leads to large interval sizes. We tackle these drawbacks by proposing a UQ method based on the predictability, computability, and stability (PCS) framework for veridical data science proposed by Yu and Kumbier. Specifically, PCS-UQ addresses model selection by using a prediction check to screen out unsuitable models. PCS-UQ then fits these screened algorithms across multiple bootstraps to assess inter-sample variability and algorithmic instability, enabling more reliable uncertainty estimates. Further, we propose a novel calibration scheme that improves local adaptivity of our prediction sets. Experiments across $17$ regression and $6$ classification datasets show that PCS-UQ achieves the desired coverage and reduces width over conformal approaches by $\approx 20\%$. Further, our local analysis shows PCS-UQ often achieves target coverage across subgroups while conformal methods fail to do so. For large deep-learning models, we propose computationally efficient approximation schemes that avoid the expensive multiple bootstrap trainings of PCS-UQ. Across three computer vision benchmarks, PCS-UQ reduces prediction set size over conformal methods by $20\%$. Theoretically, we show a modified PCS-UQ algorithm is a form of split conformal inference and achieves the desired coverage with exchangeable data.
PathM3: A Multimodal Multi-Task Multiple Instance Learning Framework for Whole Slide Image Classification and Captioning
Mar 13, 2024Abstract:In the field of computational histopathology, both whole slide images (WSIs) and diagnostic captions provide valuable insights for making diagnostic decisions. However, aligning WSIs with diagnostic captions presents a significant challenge. This difficulty arises from two main factors: 1) Gigapixel WSIs are unsuitable for direct input into deep learning models, and the redundancy and correlation among the patches demand more attention; and 2) Authentic WSI diagnostic captions are extremely limited, making it difficult to train an effective model. To overcome these obstacles, we present PathM3, a multimodal, multi-task, multiple instance learning (MIL) framework for WSI classification and captioning. PathM3 adapts a query-based transformer to effectively align WSIs with diagnostic captions. Given that histopathology visual patterns are redundantly distributed across WSIs, we aggregate each patch feature with MIL method that considers the correlations among instances. Furthermore, our PathM3 overcomes data scarcity in WSI-level captions by leveraging limited WSI diagnostic caption data in the manner of multi-task joint learning. Extensive experiments with improved classification accuracy and caption generation demonstrate the effectiveness of our method on both WSI classification and captioning task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge