Michael Tangermann
Approximate UMAP allows for high-rate online visualization of high-dimensional data streams
Apr 05, 2024



Abstract:In the BCI field, introspection and interpretation of brain signals are desired for providing feedback or to guide rapid paradigm prototyping but are challenging due to the high noise level and dimensionality of the signals. Deep neural networks are often introspected by transforming their learned feature representations into 2- or 3-dimensional subspace visualizations using projection algorithms like Uniform Manifold Approximation and Projection (UMAP). Unfortunately, these methods are computationally expensive, making the projection of data streams in real-time a non-trivial task. In this study, we introduce a novel variant of UMAP, called approximate UMAP (aUMAP). It aims at generating rapid projections for real-time introspection. To study its suitability for real-time projecting, we benchmark the methods against standard UMAP and its neural network counterpart parametric UMAP. Our results show that approximate UMAP delivers projections that replicate the projection space of standard UMAP while decreasing projection speed by an order of magnitude and maintaining the same training time.
Synthesizing EEG Signals from Event-Related Potential Paradigms with Conditional Diffusion Models
Mar 27, 2024Abstract:Data scarcity in the brain-computer interface field can be alleviated through the use of generative models, specifically diffusion models. While diffusion models have previously been successfully applied to electroencephalogram (EEG) data, existing models lack flexibility w.r.t.~sampling or require alternative representations of the EEG data. To overcome these limitations, we introduce a novel approach to conditional diffusion models that utilizes classifier-free guidance to directly generate subject-, session-, and class-specific EEG data. In addition to commonly used metrics, domain-specific metrics are employed to evaluate the specificity of the generated samples. The results indicate that the proposed model can generate EEG data that resembles real data for each subject, session, and class.
S-JEPA: towards seamless cross-dataset transfer through dynamic spatial attention
Mar 18, 2024



Abstract:Motivated by the challenge of seamless cross-dataset transfer in EEG signal processing, this article presents an exploratory study on the use of Joint Embedding Predictive Architectures (JEPAs). In recent years, self-supervised learning has emerged as a promising approach for transfer learning in various domains. However, its application to EEG signals remains largely unexplored. In this article, we introduce Signal-JEPA for representing EEG recordings which includes a novel domain-specific spatial block masking strategy and three novel architectures for downstream classification. The study is conducted on a 54~subjects dataset and the downstream performance of the models is evaluated on three different BCI paradigms: motor imagery, ERP and SSVEP. Our study provides preliminary evidence for the potential of JEPAs in EEG signal encoding. Notably, our results highlight the importance of spatial filtering for accurate downstream classification and reveal an influence of the length of the pre-training examples but not of the mask size on the downstream performance.
UMM: Unsupervised Mean-difference Maximization
Jun 20, 2023Abstract:Many brain-computer interfaces make use of brain signals that are elicited in response to a visual, auditory or tactile stimulus, so-called event-related potentials (ERPs). In visual ERP speller applications, sets of letters shown on a screen are flashed randomly, and the participant attends to the target letter they want to spell. When this letter flashes, the resulting ERP is different compared to when any other non-target letter flashes. We propose a new unsupervised approach to detect this attended letter. In each trial, for every available letter our approach makes the hypothesis that it is in fact the attended letter, and calculates the ERPs based on each of these hypotheses. We leverage the fact that only the true hypothesis produces the largest difference between the class means. Note that this unsupervised method does not require any changes to the underlying experimental paradigm and therefore can be employed in almost any ERP-based setup. To deal with limited data, we use a block-Toeplitz regularized covariance matrix that models the background activity. We implemented the proposed novel unsupervised mean-difference maximization (UMM) method and evaluated it in offline replays of brain-computer interface visual speller datasets. For a dataset that used 16 flashes per symbol per trial, UMM correctly classifies 3651 out of 3654 letters ($99.92\,\%$) across 25 participants. In another dataset with fewer and shorter trials, 7344 out of 7383 letters ($99.47\,\%$) are classified correctly across 54 participants with two sessions each. Even in more challenging datasets obtained from patients with amyotrophic lateral sclerosis ($77.86\,\%$) or when using auditory ERPs ($82.52\,\%$), the obtained classification rates obtained by UMM are competitive. In addition, UMM provides stable confidence measures which can be used to monitor convergence.
Introducing Block-Toeplitz Covariance Matrices to Remaster Linear Discriminant Analysis for Event-related Potential Brain-computer Interfaces
Feb 16, 2022

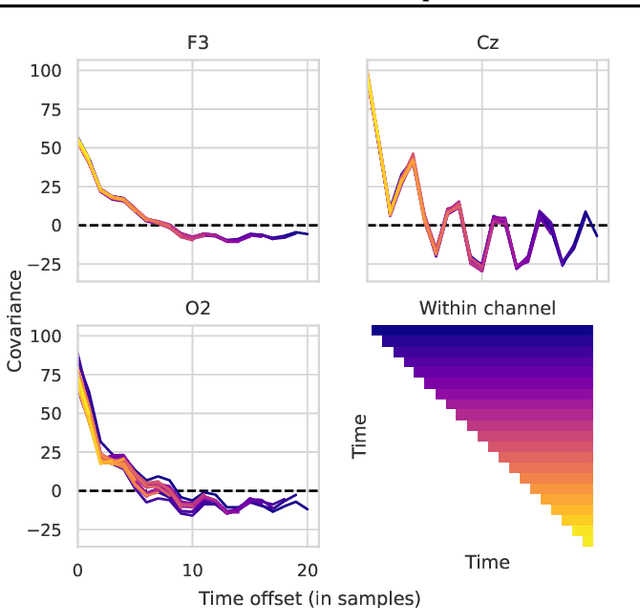
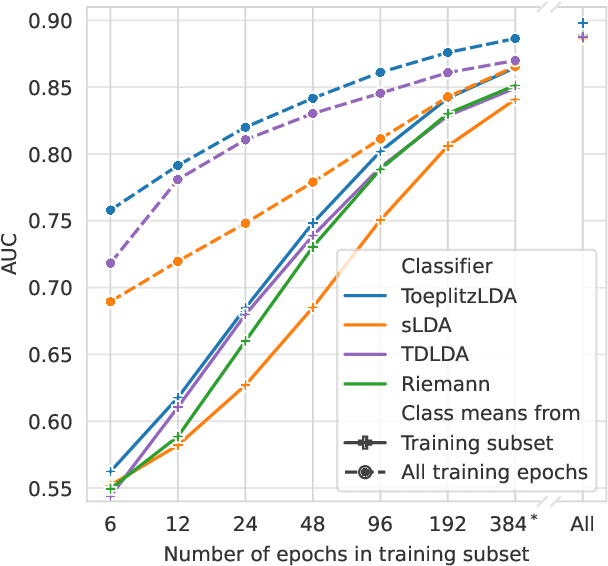
Abstract:Covariance matrices of noisy multichannel electroencephalogram time series data are hard to estimate due to high dimensionality. In brain-computer interfaces (BCI) based on event-related potentials and a linear discriminant analysis (LDA) for classification, the state of the art to address this problem is by shrinkage regularization. We propose a novel idea to tackle this problem by enforcing a block-Toeplitz structure for the covariance matrix of the LDA, which implements an assumption of signal stationarity in short time windows for each channel. On data of 213 subjects collected under 13 event-related potential BCI protocols, the resulting 'ToeplitzLDA' significantly increases the binary classification performance compared to shrinkage regularized LDA (up to 6 AUC points) and Riemannian classification approaches (up to 2 AUC points). This translates to greatly improved application level performances, as exemplified on data recorded during an unsupervised visual speller application, where spelling errors could be reduced by 81% on average for 25 subjects. Aside from lower memory and time complexity for LDA training, ToeplitzLDA proved to be almost invariant even to a twenty-fold time dimensionality enlargement, which reduces the need of expert knowledge regarding feature extraction.
Learning User Preferences for Trajectories from Brain Signals
Sep 03, 2019



Abstract:Robot motions in the presence of humans should not only be feasible and safe, but also conform to human preferences. This, however, requires user feedback on the robot's behavior. In this work, we propose a novel approach to leverage the user's brain signals as a feedback modality in order to decode the judgment of robot trajectories and rank them according to the user's preferences. We show that brain signals measured using electroencephalography during observation of a robotic arm's trajectory as well as in response to preference statements are informative regarding the user's preference. Furthermore, we demonstrate that user feedback from brain signals can be used to reliably infer pairwise trajectory preferences as well as to retrieve the preferred observed trajectories of the user with a performance comparable to explicit behavioral feedback.
Deep learning with convolutional neural networks for EEG decoding and visualization
Jun 08, 2018



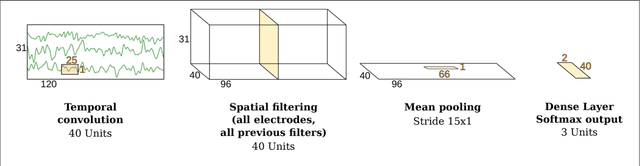
Abstract:PLEASE READ AND CITE THE REVISED VERSION at Human Brain Mapping: http://onlinelibrary.wiley.com/doi/10.1002/hbm.23730/full Code available here: https://github.com/robintibor/braindecode
Method to assess the functional role of noisy brain signals by mining envelope dynamics
Apr 27, 2018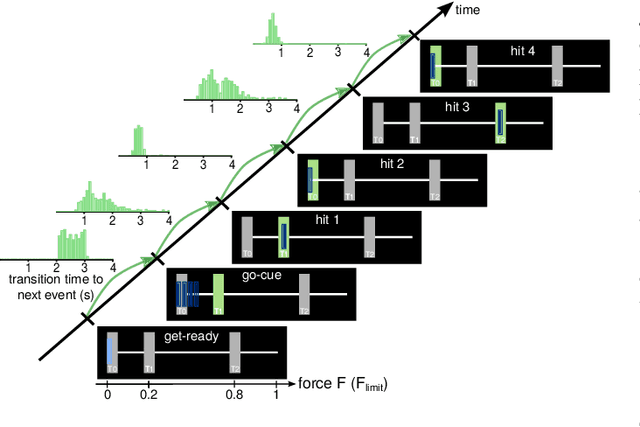
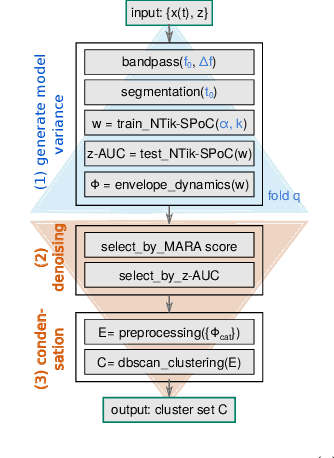
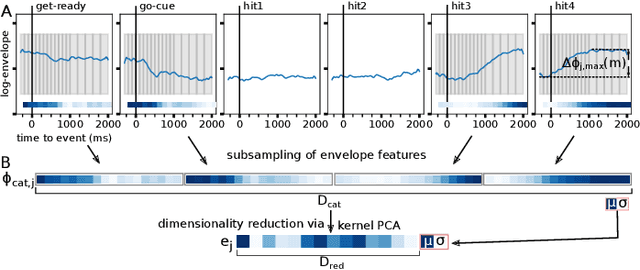
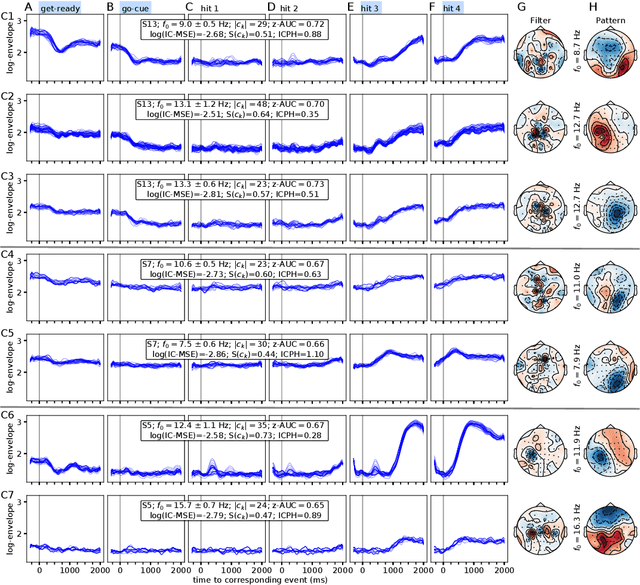
Abstract:Data-driven spatial filtering approaches are commonly used to assess rhythmic brain activity from multichannel recordings such as electroencephalography (EEG). As spatial filter estimation is prone to noise, non-stationarity effects and limited data, a high model variability induced by slight changes of, e.g., involved hyperparameters is generally encountered. These aspects challenge the assessment of functionally relevant features which are of special importance in closed-loop applications as, e.g., in the field of rehabilitation. We propose a data-driven method to identify groups of reliable and functionally relevant oscillatory components computed by a spatial filtering approach. Therefore, we initially embrace the variability of decoding models in a large configuration space before condensing information by density-based clustering of components' functional signatures. Exemplified for a hand force task with rich within-trial structure, the approach was evaluated on EEG data of 18 healthy subjects. We found that functional characteristics of single components are revealed by distinct temporal dynamics of their event-related power changes. Based on a within-subject analysis, our clustering revealed seven groups of homogeneous envelope dynamics on average. To support introspection by practitioners, we provide a set of metrics to characterize and validate single clusterings. We show that identified clusters contain components of strictly confined frequency ranges, dominated by the alpha and beta band. Our method is applicable to any spatial filtering algorithm. Despite high model variability, it allows capturing and monitoring relevant oscillatory features. We foresee its application in closed-loop applications such as brain-computer interface based protocols in stroke rehabilitation.
Post-hoc labeling of arbitrary EEG recordings for data-efficient evaluation of neural decoding methods
Nov 22, 2017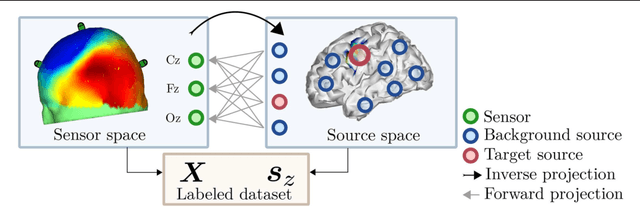
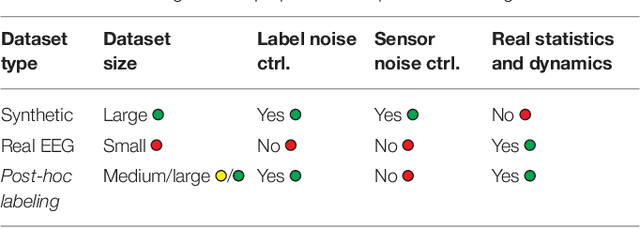
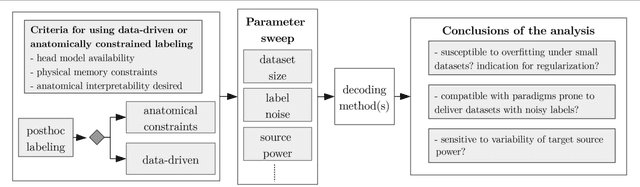
Abstract:Many cognitive, sensory and motor processes have correlates in oscillatory neural sources, which are embedded as a subspace into the recorded brain signals. Decoding such processes from noisy magnetoencephalogram/electroencephalogram (M/EEG) signals usually requires the use of data-driven analysis methods. The objective evaluation of such decoding algorithms on experimental raw signals, however, is a challenge: the amount of available M/EEG data typically is limited, labels can be unreliable, and raw signals often are contaminated with artifacts. The latter is specifically problematic, if the artifacts stem from behavioral confounds of the oscillatory neural processes of interest. To overcome some of these problems, simulation frameworks have been introduced for benchmarking decoding methods. Generating artificial brain signals, however, most simulation frameworks make strong and partially unrealistic assumptions about brain activity, which limits the generalization of obtained results to real-world conditions. In the present contribution, we thrive to remove many shortcomings of current simulation frameworks and propose a versatile alternative, that allows for objective evaluation and benchmarking of novel data-driven decoding methods for neural signals. Its central idea is to utilize post-hoc labelings of arbitrary M/EEG recordings. This strategy makes it paradigm-agnostic and allows to generate comparatively large datasets with noiseless labels. Source code and data of the novel simulation approach are made available for facilitating its adoption.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge