Andreas Meinel
Method to assess the functional role of noisy brain signals by mining envelope dynamics
Apr 27, 2018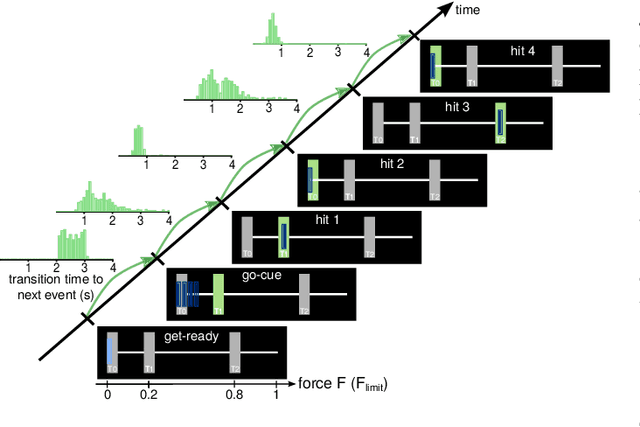
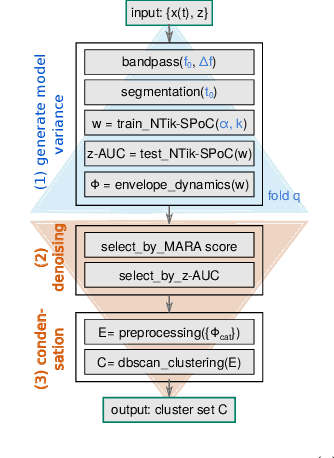
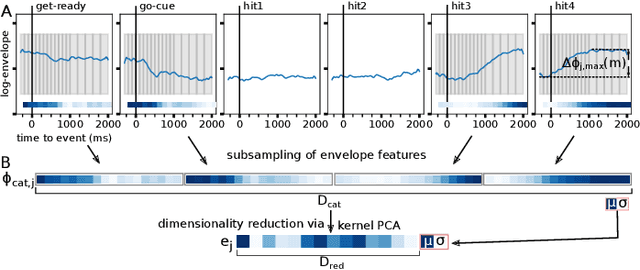
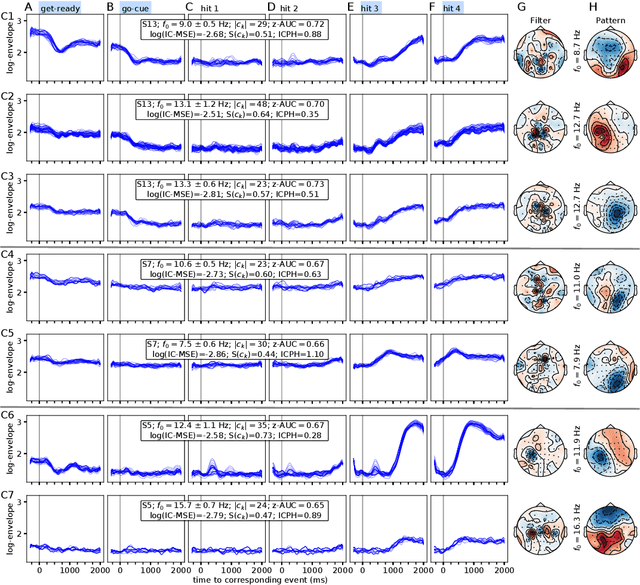
Abstract:Data-driven spatial filtering approaches are commonly used to assess rhythmic brain activity from multichannel recordings such as electroencephalography (EEG). As spatial filter estimation is prone to noise, non-stationarity effects and limited data, a high model variability induced by slight changes of, e.g., involved hyperparameters is generally encountered. These aspects challenge the assessment of functionally relevant features which are of special importance in closed-loop applications as, e.g., in the field of rehabilitation. We propose a data-driven method to identify groups of reliable and functionally relevant oscillatory components computed by a spatial filtering approach. Therefore, we initially embrace the variability of decoding models in a large configuration space before condensing information by density-based clustering of components' functional signatures. Exemplified for a hand force task with rich within-trial structure, the approach was evaluated on EEG data of 18 healthy subjects. We found that functional characteristics of single components are revealed by distinct temporal dynamics of their event-related power changes. Based on a within-subject analysis, our clustering revealed seven groups of homogeneous envelope dynamics on average. To support introspection by practitioners, we provide a set of metrics to characterize and validate single clusterings. We show that identified clusters contain components of strictly confined frequency ranges, dominated by the alpha and beta band. Our method is applicable to any spatial filtering algorithm. Despite high model variability, it allows capturing and monitoring relevant oscillatory features. We foresee its application in closed-loop applications such as brain-computer interface based protocols in stroke rehabilitation.
Post-hoc labeling of arbitrary EEG recordings for data-efficient evaluation of neural decoding methods
Nov 22, 2017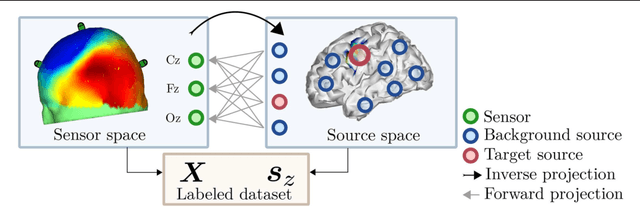
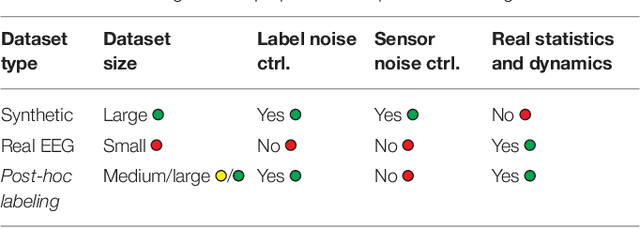
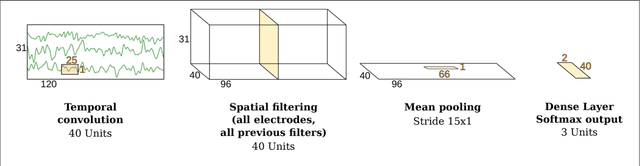
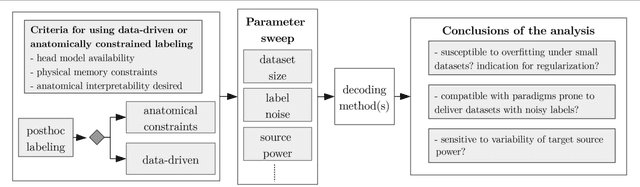
Abstract:Many cognitive, sensory and motor processes have correlates in oscillatory neural sources, which are embedded as a subspace into the recorded brain signals. Decoding such processes from noisy magnetoencephalogram/electroencephalogram (M/EEG) signals usually requires the use of data-driven analysis methods. The objective evaluation of such decoding algorithms on experimental raw signals, however, is a challenge: the amount of available M/EEG data typically is limited, labels can be unreliable, and raw signals often are contaminated with artifacts. The latter is specifically problematic, if the artifacts stem from behavioral confounds of the oscillatory neural processes of interest. To overcome some of these problems, simulation frameworks have been introduced for benchmarking decoding methods. Generating artificial brain signals, however, most simulation frameworks make strong and partially unrealistic assumptions about brain activity, which limits the generalization of obtained results to real-world conditions. In the present contribution, we thrive to remove many shortcomings of current simulation frameworks and propose a versatile alternative, that allows for objective evaluation and benchmarking of novel data-driven decoding methods for neural signals. Its central idea is to utilize post-hoc labelings of arbitrary M/EEG recordings. This strategy makes it paradigm-agnostic and allows to generate comparatively large datasets with noiseless labels. Source code and data of the novel simulation approach are made available for facilitating its adoption.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge