Michael Quail
Image2Flow: A hybrid image and graph convolutional neural network for rapid patient-specific pulmonary artery segmentation and CFD flow field calculation from 3D cardiac MRI data
Feb 28, 2024Abstract:Computational fluid dynamics (CFD) can be used for evaluation of hemodynamics. However, its routine use is limited by labor-intensive manual segmentation, CFD mesh creation, and time-consuming simulation. This study aims to train a deep learning model to both generate patient-specific volume-meshes of the pulmonary artery from 3D cardiac MRI data and directly estimate CFD flow fields. This study used 135 3D cardiac MRIs from both a public and private dataset. The pulmonary arteries in the MRIs were manually segmented and converted into volume-meshes. CFD simulations were performed on ground truth meshes and interpolated onto point-point correspondent meshes to create the ground truth dataset. The dataset was split 85/10/15 for training, validation and testing. Image2Flow, a hybrid image and graph convolutional neural network, was trained to transform a pulmonary artery template to patient-specific anatomy and CFD values. Image2Flow was evaluated in terms of segmentation and accuracy of CFD predicted was assessed using node-wise comparisons. Centerline comparisons of Image2Flow and CFD simulations performed using machine learning segmentation were also performed. Image2Flow achieved excellent segmentation accuracy with a median Dice score of 0.9 (IQR: 0.86-0.92). The median node-wise normalized absolute error for pressure and velocity magnitude was 11.98% (IQR: 9.44-17.90%) and 8.06% (IQR: 7.54-10.41), respectively. Centerline analysis showed no significant difference between the Image2Flow and conventional CFD simulated on machine learning-generated volume-meshes. This proof-of-concept study has shown it is possible to simultaneously perform patient specific volume-mesh based segmentation and pressure and flow field estimation. Image2Flow completes segmentation and CFD in ~205ms, which ~7000 times faster than manual methods, making it more feasible in a clinical environment.
Deep Learning Pipeline for Preprocessing and Segmenting Cardiac Magnetic Resonance of Single Ventricle Patients from an Image Registry
Mar 21, 2023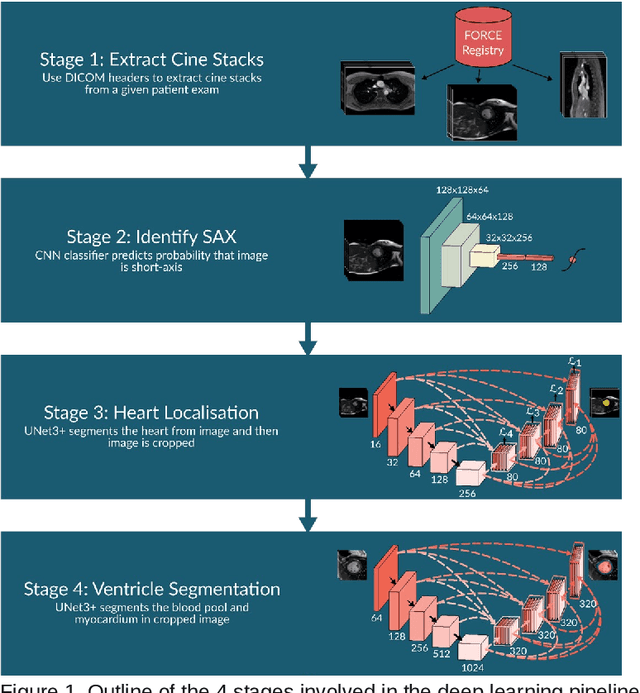
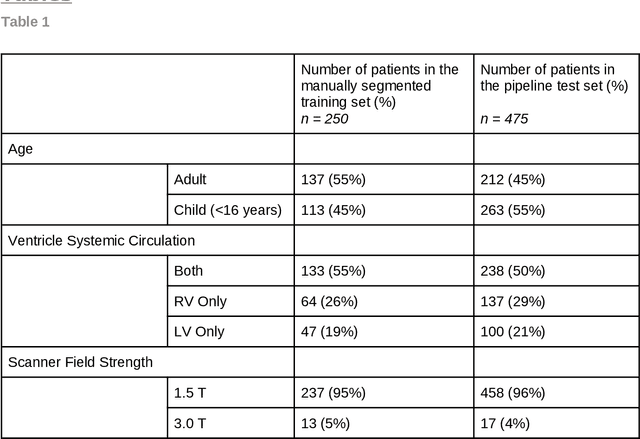
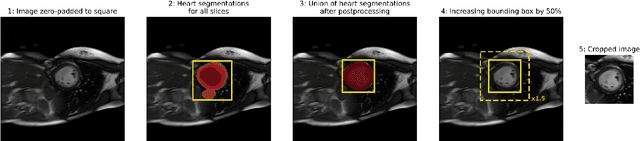
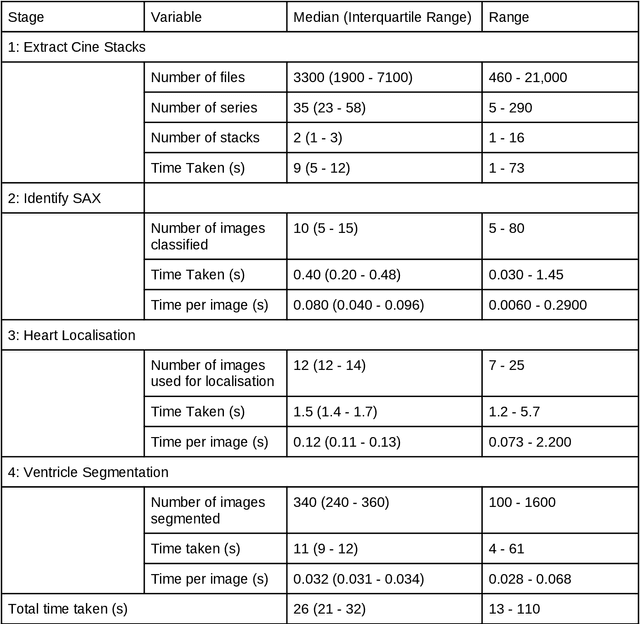
Abstract:Purpose: To develop and evaluate an end-to-end deep learning pipeline for segmentation and analysis of cardiac magnetic resonance images to provide core-lab processing for a multi-centre registry of Fontan patients. Materials and Methods: This retrospective study used training (n = 175), validation (n = 25) and testing (n = 50) cardiac magnetic resonance image exams collected from 13 institutions in the UK, US and Canada. The data was used to train and evaluate a pipeline containing three deep-learning models. The pipeline's performance was assessed on the Dice and IoU score between the automated and reference standard manual segmentation. Cardiac function values were calculated from both the automated and manual segmentation and evaluated using Bland-Altman analysis and paired t-tests. The overall pipeline was further evaluated qualitatively on 475 unseen patient exams. Results: For the 50 testing dataset, the pipeline achieved a median Dice score of 0.91 (0.89-0.94) for end-diastolic volume, 0.86 (0.82-0.89) for end-systolic volume, and 0.74 (0.70-0.77) for myocardial mass. The deep learning-derived end-diastolic volume, end-systolic volume, myocardial mass, stroke volume and ejection fraction had no statistical difference compared to the same values derived from manual segmentation with p values all greater than 0.05. For the 475 unseen patient exams, the pipeline achieved 68% adequate segmentation in both systole and diastole, 26% needed minor adjustments in either systole or diastole, 5% needed major adjustments, and the cropping model only failed in 0.4%. Conclusion: Deep learning pipeline can provide standardised 'core-lab' segmentation for Fontan patients. This pipeline can now be applied to the >4500 cardiac magnetic resonance exams currently in the FORCE registry as well as any new patients that are recruited.
Rapid Whole-Heart CMR with Single Volume Super-resolution
Dec 22, 2019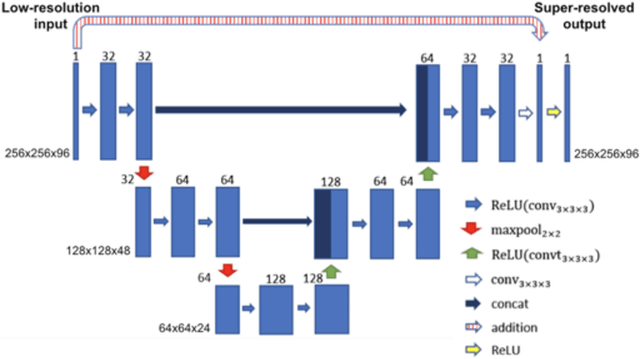
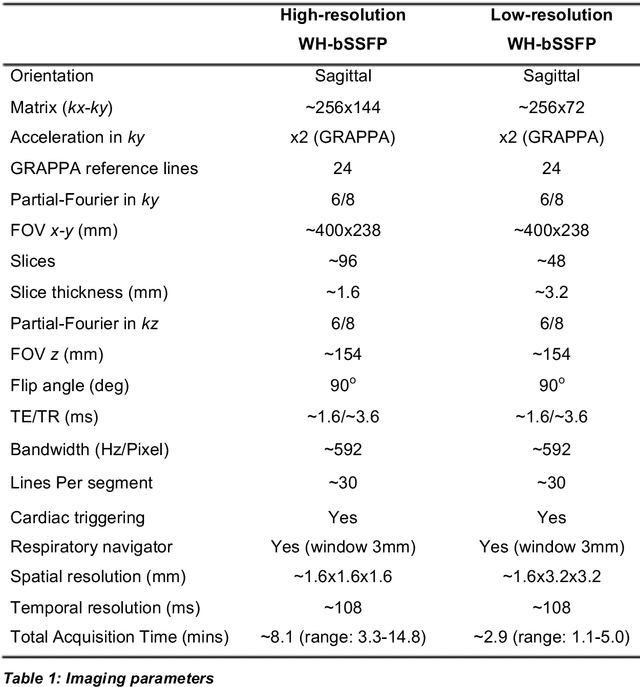
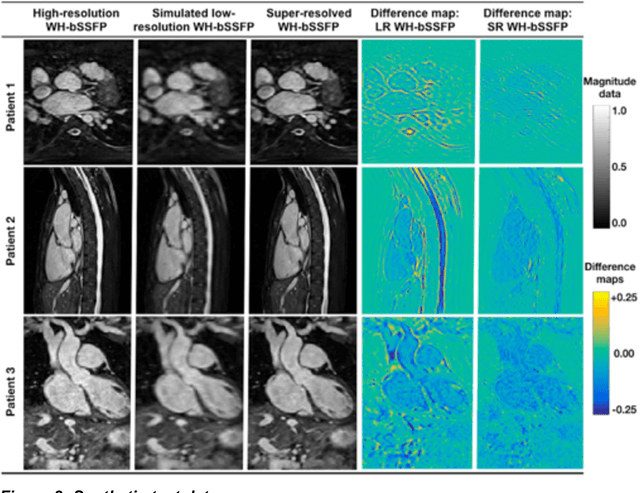
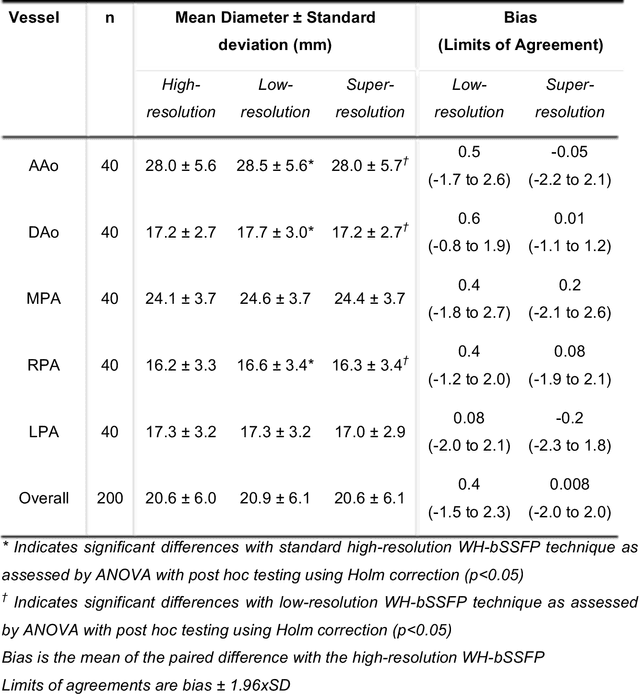
Abstract:Background: Three-dimensional, whole heart, balanced steady state free precession (WH-bSSFP) sequences provide delineation of intra-cardiac and vascular anatomy. However, they have long acquisition times. Here, we propose significant speed ups using a deep learning single volume super resolution reconstruction, to recover high resolution features from rapidly acquired low resolution WH-bSSFP images. Methods: A 3D residual U-Net was trained using synthetic data, created from a library of high-resolution WH-bSSFP images by simulating 0.5 slice resolution and 0.5 phase resolution. The trained network was validated with synthetic test data, as well as prospective low-resolution data. Results: Synthetic low-resolution data had significantly better image quality after super-resolution reconstruction. Qualitative image scores showed super-resolved images had better edge sharpness, fewer residual artefacts and less image distortion than low-resolution images, with similar scores to high-resolution data. Quantitative image scores showed super-resolved images had significantly better edge sharpness than low-resolution or high-resolution images, with significantly better signal-to-noise ratio than high-resolution data. Vessel diameters measurements showed over-estimation in the low-resolution measurements, compared to the high-resolution data. No significant differences and no bias was found in the super-resolution measurements. Conclusion: This paper demonstrates the potential of using a residual U-Net for super-resolution reconstruction of rapidly acquired low-resolution whole heart bSSFP data within a clinical setting. The resulting network can be applied very quickly, making these techniques particularly appealing within busy clinical workflow. Thus, we believe that this technique may help speed up whole heart CMR in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge