Gabriel F. Miller
MultiFlow: A unified deep learning framework for multi-vessel classification, segmentation and clustering of phase-contrast MRI validated on a multi-site single ventricle patient cohort
Feb 17, 2025



Abstract:This study presents a unified deep learning (DL) framework, MultiFlowSeg, for classification and segmentation of velocity-encoded phase-contrast magnetic resonance imaging data, and MultiFlowDTC for temporal clustering of flow phenotypes. Applied to the FORCE registry of Fontan procedure patients, MultiFlowSeg achieved 100% classification accuracy for the aorta, SVC, and IVC, and 94% for the LPA and RPA. It demonstrated robust segmentation with a median Dice score of 0.91 (IQR: 0.86-0.93). The automated pipeline processed registry data, achieving high segmentation success despite challenges like poor image quality and dextrocardia. Temporal clustering identified five distinct patient subgroups, with significant differences in clinical outcomes, including ejection fraction, exercise tolerance, liver disease, and mortality. These results demonstrate the potential of combining DL and time-varying flow data for improved CHD prognosis and personalized care.
Deep Learning Pipeline for Preprocessing and Segmenting Cardiac Magnetic Resonance of Single Ventricle Patients from an Image Registry
Mar 21, 2023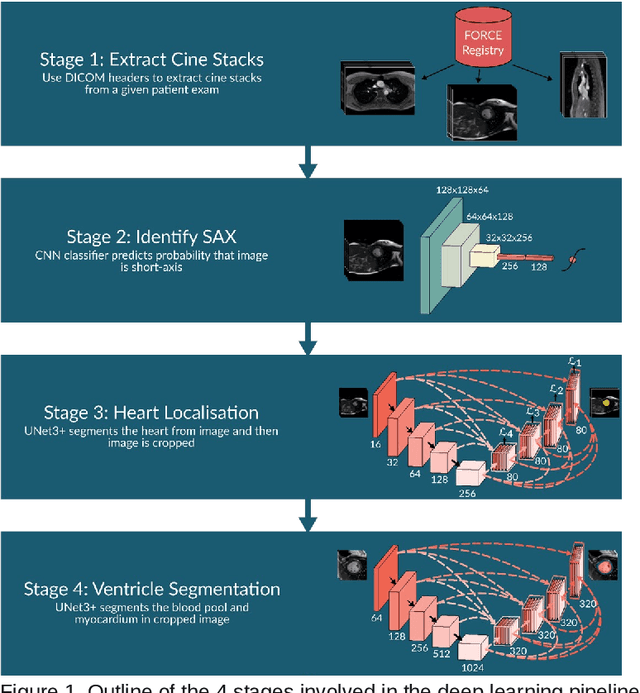
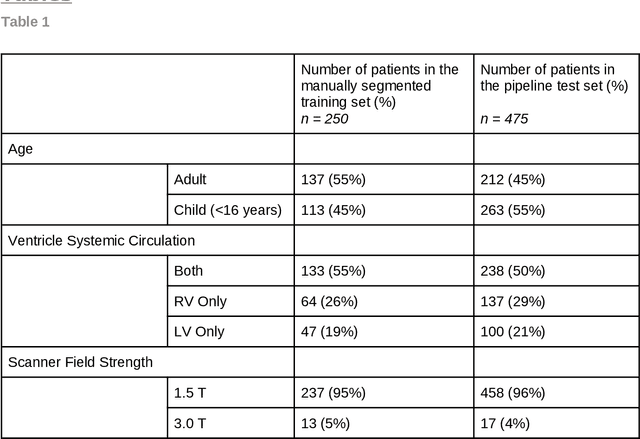
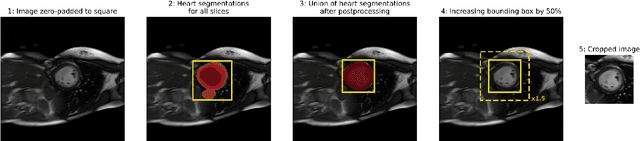
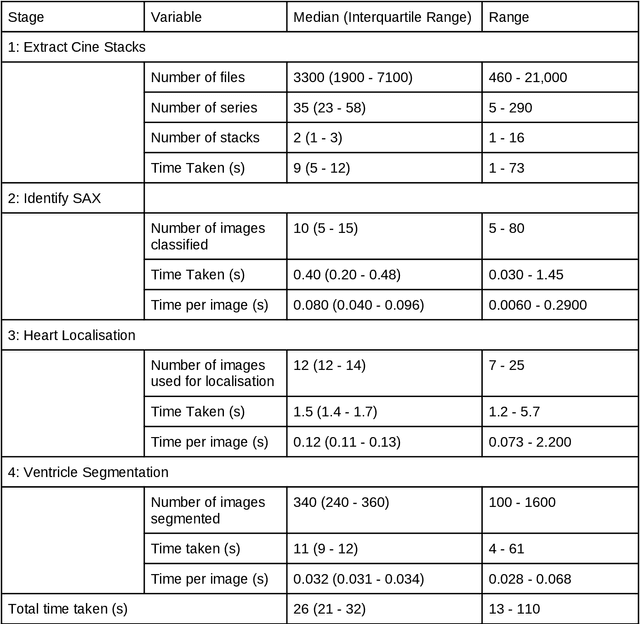
Abstract:Purpose: To develop and evaluate an end-to-end deep learning pipeline for segmentation and analysis of cardiac magnetic resonance images to provide core-lab processing for a multi-centre registry of Fontan patients. Materials and Methods: This retrospective study used training (n = 175), validation (n = 25) and testing (n = 50) cardiac magnetic resonance image exams collected from 13 institutions in the UK, US and Canada. The data was used to train and evaluate a pipeline containing three deep-learning models. The pipeline's performance was assessed on the Dice and IoU score between the automated and reference standard manual segmentation. Cardiac function values were calculated from both the automated and manual segmentation and evaluated using Bland-Altman analysis and paired t-tests. The overall pipeline was further evaluated qualitatively on 475 unseen patient exams. Results: For the 50 testing dataset, the pipeline achieved a median Dice score of 0.91 (0.89-0.94) for end-diastolic volume, 0.86 (0.82-0.89) for end-systolic volume, and 0.74 (0.70-0.77) for myocardial mass. The deep learning-derived end-diastolic volume, end-systolic volume, myocardial mass, stroke volume and ejection fraction had no statistical difference compared to the same values derived from manual segmentation with p values all greater than 0.05. For the 475 unseen patient exams, the pipeline achieved 68% adequate segmentation in both systole and diastole, 26% needed minor adjustments in either systole or diastole, 5% needed major adjustments, and the cropping model only failed in 0.4%. Conclusion: Deep learning pipeline can provide standardised 'core-lab' segmentation for Fontan patients. This pipeline can now be applied to the >4500 cardiac magnetic resonance exams currently in the FORCE registry as well as any new patients that are recruited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge