Mark A. Fogel
Deep Learning Pipeline for Preprocessing and Segmenting Cardiac Magnetic Resonance of Single Ventricle Patients from an Image Registry
Mar 21, 2023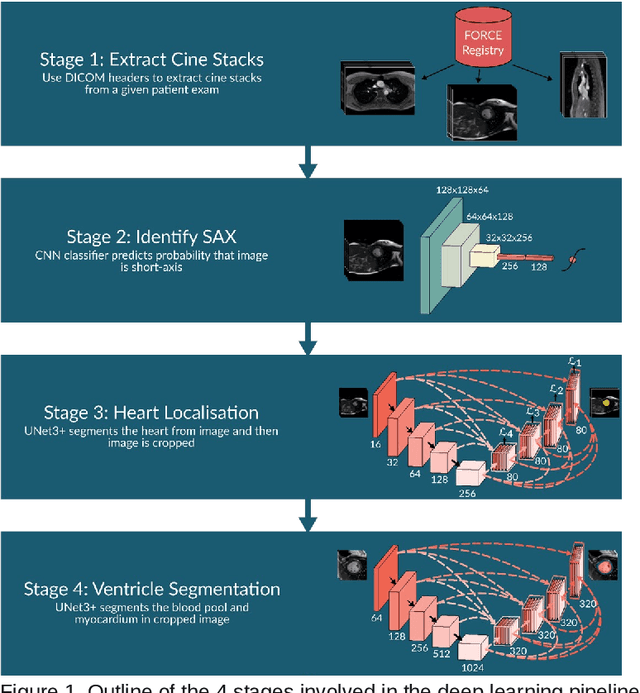
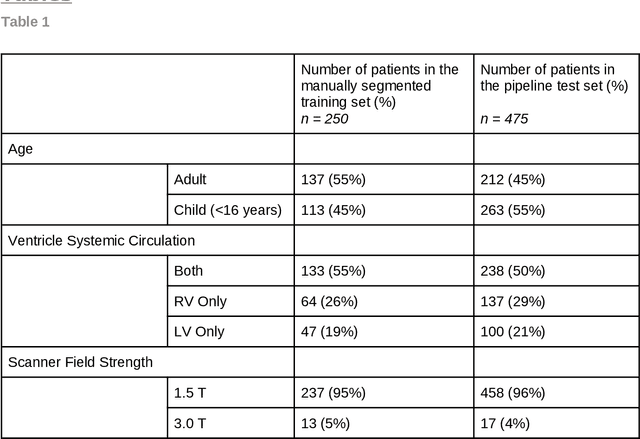
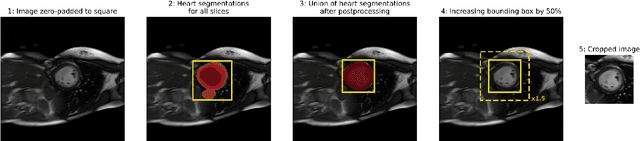
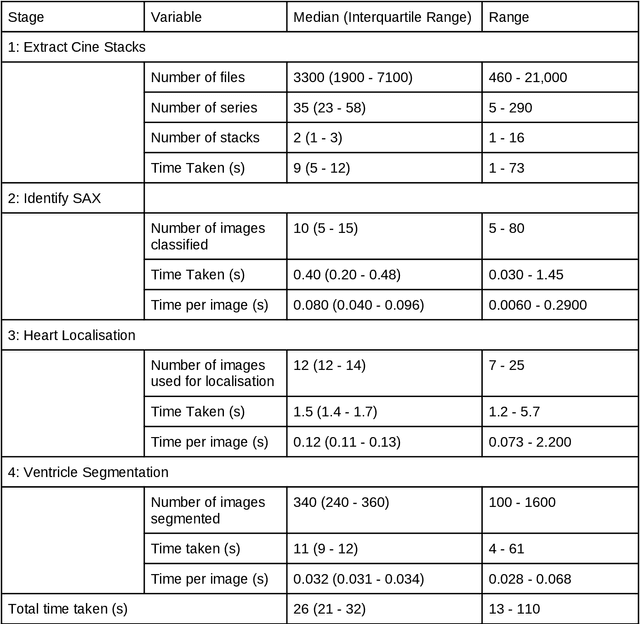
Abstract:Purpose: To develop and evaluate an end-to-end deep learning pipeline for segmentation and analysis of cardiac magnetic resonance images to provide core-lab processing for a multi-centre registry of Fontan patients. Materials and Methods: This retrospective study used training (n = 175), validation (n = 25) and testing (n = 50) cardiac magnetic resonance image exams collected from 13 institutions in the UK, US and Canada. The data was used to train and evaluate a pipeline containing three deep-learning models. The pipeline's performance was assessed on the Dice and IoU score between the automated and reference standard manual segmentation. Cardiac function values were calculated from both the automated and manual segmentation and evaluated using Bland-Altman analysis and paired t-tests. The overall pipeline was further evaluated qualitatively on 475 unseen patient exams. Results: For the 50 testing dataset, the pipeline achieved a median Dice score of 0.91 (0.89-0.94) for end-diastolic volume, 0.86 (0.82-0.89) for end-systolic volume, and 0.74 (0.70-0.77) for myocardial mass. The deep learning-derived end-diastolic volume, end-systolic volume, myocardial mass, stroke volume and ejection fraction had no statistical difference compared to the same values derived from manual segmentation with p values all greater than 0.05. For the 475 unseen patient exams, the pipeline achieved 68% adequate segmentation in both systole and diastole, 26% needed minor adjustments in either systole or diastole, 5% needed major adjustments, and the cropping model only failed in 0.4%. Conclusion: Deep learning pipeline can provide standardised 'core-lab' segmentation for Fontan patients. This pipeline can now be applied to the >4500 cardiac magnetic resonance exams currently in the FORCE registry as well as any new patients that are recruited.
Free-running SIMilarity-Based Angiography (SIMBA) for simplified anatomical MR imaging of the heart
Jul 13, 2020



Abstract:Purpose: Whole-heart MRA techniques typically target pre-determined motion states and address cardiac and respiratory dynamics independently. We propose a novel fast reconstruction algorithm, applicable to ungated free-running sequences, that leverages inherent similarities in the acquired data to avoid such physiological constraints. Theory and Methods: The proposed SIMilarity-Based Angiography (SIMBA) method clusters the continuously acquired k-space data in order to find a motion-consistent subset that can be reconstructed into a motion-suppressed whole-heart MRA. Free-running 3D radial datasets from six ferumoxytol-enhanced scans of pediatric cardiac patients and twelve non-contrast scans of healthy volunteers were reconstructed with a non-motion-suppressed regridding of all the acquired data (All Data), our proposed SIMBA method, and a previously published free-running framework (FRF) that uses cardiac and respiratory self-gating and compressed sensing. Images were compared for blood-myocardium interface sharpness, contrast ratio, and visibility of coronary artery ostia. Results: Both the fast SIMBA reconstruction (~20s) and the FRF provided significantly higher blood-myocardium sharpness than All Data (P<0.001). No significant difference was observed among the former two. Significantly higher blood-myocardium contrast ratio was obtained with SIMBA compared to All Data and FRF (P<0.01). More coronary ostia could be visualized with both SIMBA and FRF than with All Data (All Data: 4/36, SIMBA: 30/36, FRF: 33/36, both P<0.001) but no significant difference was found between the first two. Conclusion: The combination of free-running sequences and the fast SIMBA reconstruction, which operates without a priori assumptions related to physiological motion, forms a simple workflow for obtaining whole-heart MRA with sharp anatomical structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge