Silvia Schievano
Image2Flow: A hybrid image and graph convolutional neural network for rapid patient-specific pulmonary artery segmentation and CFD flow field calculation from 3D cardiac MRI data
Feb 28, 2024Abstract:Computational fluid dynamics (CFD) can be used for evaluation of hemodynamics. However, its routine use is limited by labor-intensive manual segmentation, CFD mesh creation, and time-consuming simulation. This study aims to train a deep learning model to both generate patient-specific volume-meshes of the pulmonary artery from 3D cardiac MRI data and directly estimate CFD flow fields. This study used 135 3D cardiac MRIs from both a public and private dataset. The pulmonary arteries in the MRIs were manually segmented and converted into volume-meshes. CFD simulations were performed on ground truth meshes and interpolated onto point-point correspondent meshes to create the ground truth dataset. The dataset was split 85/10/15 for training, validation and testing. Image2Flow, a hybrid image and graph convolutional neural network, was trained to transform a pulmonary artery template to patient-specific anatomy and CFD values. Image2Flow was evaluated in terms of segmentation and accuracy of CFD predicted was assessed using node-wise comparisons. Centerline comparisons of Image2Flow and CFD simulations performed using machine learning segmentation were also performed. Image2Flow achieved excellent segmentation accuracy with a median Dice score of 0.9 (IQR: 0.86-0.92). The median node-wise normalized absolute error for pressure and velocity magnitude was 11.98% (IQR: 9.44-17.90%) and 8.06% (IQR: 7.54-10.41), respectively. Centerline analysis showed no significant difference between the Image2Flow and conventional CFD simulated on machine learning-generated volume-meshes. This proof-of-concept study has shown it is possible to simultaneously perform patient specific volume-mesh based segmentation and pressure and flow field estimation. Image2Flow completes segmentation and CFD in ~205ms, which ~7000 times faster than manual methods, making it more feasible in a clinical environment.
Latent Disentanglement in Mesh Variational Autoencoders Improves the Diagnosis of Craniofacial Syndromes and Aids Surgical Planning
Sep 05, 2023Abstract:The use of deep learning to undertake shape analysis of the complexities of the human head holds great promise. However, there have traditionally been a number of barriers to accurate modelling, especially when operating on both a global and local level. In this work, we will discuss the application of the Swap Disentangled Variational Autoencoder (SD-VAE) with relevance to Crouzon, Apert and Muenke syndromes. Although syndrome classification is performed on the entire mesh, it is also possible, for the first time, to analyse the influence of each region of the head on the syndromic phenotype. By manipulating specific parameters of the generative model, and producing procedure-specific new shapes, it is also possible to simulate the outcome of a range of craniofacial surgical procedures. This opens new avenues to advance diagnosis, aids surgical planning and allows for the objective evaluation of surgical outcomes.
Deep neural networks for fast acquisition of aortic 3D pressure and velocity flow fields
Aug 25, 2022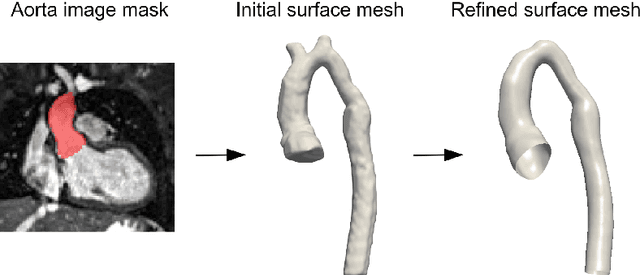
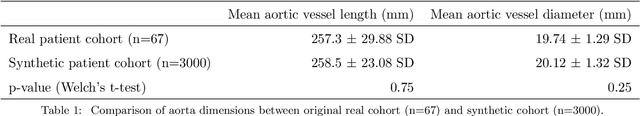
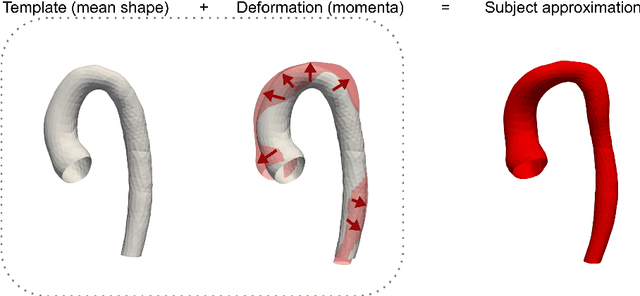
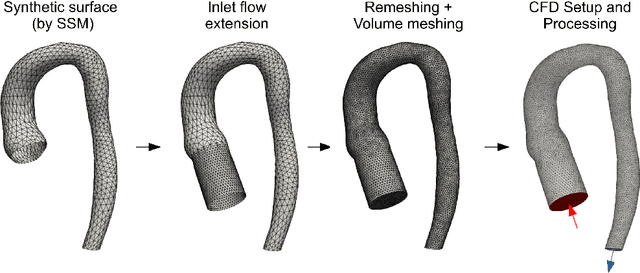
Abstract:Computational fluid dynamics (CFD) can be used to simulate vascular haemodynamics and analyse potential treatment options. CFD has shown to be beneficial in improving patient outcomes. However, the implementation of CFD for routine clinical use is yet to be realised. Barriers for CFD include high computational resources, specialist experience needed for designing simulation set-ups, and long processing times. The aim of this study was to explore the use of machine learning (ML) to replicate conventional aortic CFD with automatic and fast regression models. Data used to train/test the model comprised of 3,000 CFD simulations performed on synthetically generated 3D aortic shapes. These subjects were generated from a statistical shape model (SSM) built on real patient-specific aortas (N=67). Inference performed on 200 test shapes resulted in average errors of 6.01% +/-3.12 SD and 3.99% +/-0.93 SD for pressure and velocity, respectively. Our ML-based models performed CFD in ~0.075 seconds (4,000x faster than the solver). This study shows that results from conventional vascular CFD can be reproduced using ML at a much faster rate, in an automatic process, and with high accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge