Michael Kirby
A Granular Grassmannian Clustering Framework via the Schubert Variety of Best Fit
Dec 29, 2025Abstract:In many classification and clustering tasks, it is useful to compute a geometric representative for a dataset or a cluster, such as a mean or median. When datasets are represented by subspaces, these representatives become points on the Grassmann or flag manifold, with distances induced by their geometry, often via principal angles. We introduce a subspace clustering algorithm that replaces subspace means with a trainable prototype defined as a Schubert Variety of Best Fit (SVBF) - a subspace that comes as close as possible to intersecting each cluster member in at least one fixed direction. Integrated in the Linde-Buzo-Grey (LBG) pipeline, this SVBF-LBG scheme yields improved cluster purity on synthetic, image, spectral, and video action data, while retaining the mathematical structure required for downstream analysis.
Fine-tuning foundational models to code diagnoses from veterinary health records
Oct 19, 2024Abstract:Veterinary medical records represent a large data resource for application to veterinary and One Health clinical research efforts. Use of the data is limited by interoperability challenges including inconsistent data formats and data siloing. Clinical coding using standardized medical terminologies enhances the quality of medical records and facilitates their interoperability with veterinary and human health records from other sites. Previous studies, such as DeepTag and VetTag, evaluated the application of Natural Language Processing (NLP) to automate veterinary diagnosis coding, employing long short-term memory (LSTM) and transformer models to infer a subset of Systemized Nomenclature of Medicine - Clinical Terms (SNOMED-CT) diagnosis codes from free-text clinical notes. This study expands on these efforts by incorporating all 7,739 distinct SNOMED-CT diagnosis codes recognized by the Colorado State University (CSU) Veterinary Teaching Hospital (VTH) and by leveraging the increasing availability of pre-trained large language models (LLMs). Ten freely-available pre-trained LLMs were fine-tuned on the free-text notes from 246,473 manually-coded veterinary patient visits included in the CSU VTH's electronic health records (EHRs), which resulted in superior performance relative to previous efforts. The most accurate results were obtained when expansive labeled data were used to fine-tune relatively large clinical LLMs, but the study also showed that comparable results can be obtained using more limited resources and non-clinical LLMs. The results of this study contribute to the improvement of the quality of veterinary EHRs by investigating accessible methods for automated coding and support both animal and human health research by paving the way for more integrated and comprehensive health databases that span species and institutions.
Optimizing Sparse Generalized Singular Vectors for Feature Selection in Proximal Support Vector Machines with Application to Breast and Ovarian Cancer Detection
Oct 04, 2024
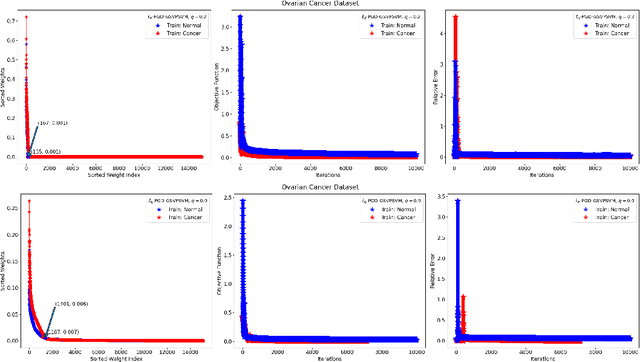
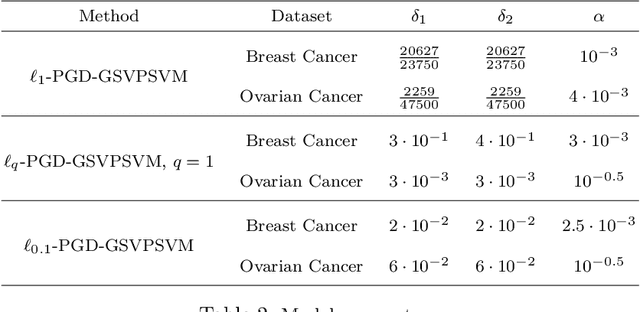
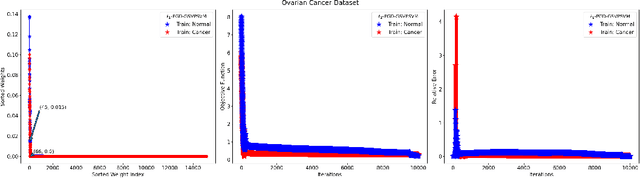
Abstract:This paper presents approaches to compute sparse solutions of Generalized Singular Value Problem (GSVP). The GSVP is regularized by $\ell_1$-norm and $\ell_q$-penalty for $0<q<1$, resulting in the $\ell_1$-GSVP and $\ell_q$-GSVP formulations. The solutions of these problems are determined by applying the proximal gradient descent algorithm with a fixed step size. The inherent sparsity levels within the computed solutions are exploited for feature selection, and subsequently, binary classification with non-parallel Support Vector Machines (SVM). For our feature selection task, SVM is integrated into the $\ell_1$-GSVP and $\ell_q$-GSVP frameworks to derive the $\ell_1$-GSVPSVM and $\ell_q$-GSVPSVM variants. Machine learning applications to cancer detection are considered. We remarkably report near-to-perfect balanced accuracy across breast and ovarian cancer datasets using a few selected features.
A Multi-Domain Multi-Task Approach for Feature Selection from Bulk RNA Datasets
May 04, 2024Abstract:In this paper a multi-domain multi-task algorithm for feature selection in bulk RNAseq data is proposed. Two datasets are investigated arising from mouse host immune response to Salmonella infection. Data is collected from several strains of collaborative cross mice. Samples from the spleen and liver serve as the two domains. Several machine learning experiments are conducted and the small subset of discriminative across domains features have been extracted in each case. The algorithm proves viable and underlines the benefits of across domain feature selection by extracting new subset of discriminative features which couldn't be extracted only by one-domain approach.
ReLU Neural Networks, Polyhedral Decompositions, and Persistent Homolog
Jun 30, 2023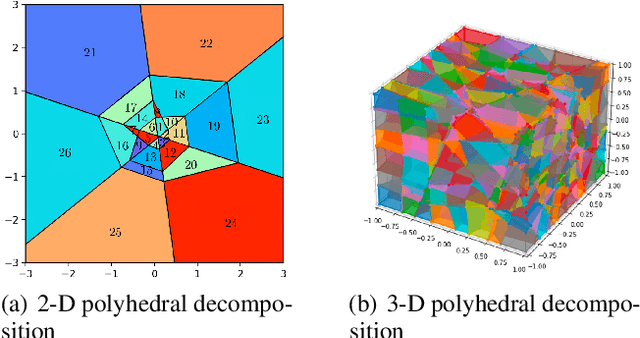
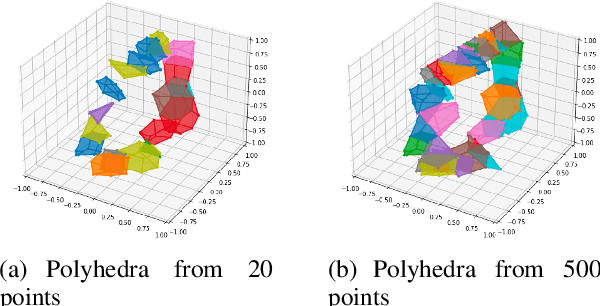
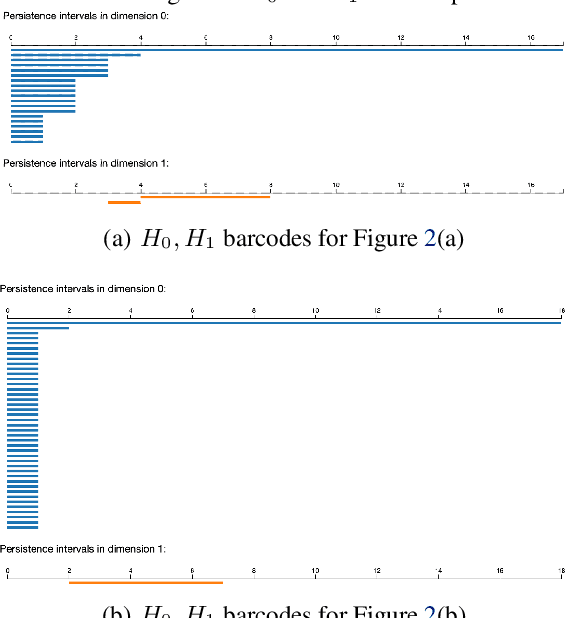

Abstract:A ReLU neural network leads to a finite polyhedral decomposition of input space and a corresponding finite dual graph. We show that while this dual graph is a coarse quantization of input space, it is sufficiently robust that it can be combined with persistent homology to detect homological signals of manifolds in the input space from samples. This property holds for a variety of networks trained for a wide range of purposes that have nothing to do with this topological application. We found this feature to be surprising and interesting; we hope it will also be useful.
Sparse Linear Centroid-Encoder: A Convex Method for Feature Selection
Jun 09, 2023Abstract:We present a novel feature selection technique, Sparse Linear Centroid-Encoder (SLCE). The algorithm uses a linear transformation to reconstruct a point as its class centroid and, at the same time, uses the $\ell_1$-norm penalty to filter out unnecessary features from the input data. The original formulation of the optimization problem is nonconvex, but we propose a two-step approach, where each step is convex. In the first step, we solve the linear Centroid-Encoder, a convex optimization problem over a matrix $A$. In the second step, we only search for a sparse solution over a diagonal matrix $B$ while keeping $A$ fixed. Unlike other linear methods, e.g., Sparse Support Vector Machines and Lasso, Sparse Linear Centroid-Encoder uses a single model for multi-class data. We present an in-depth empirical analysis of the proposed model and show that it promotes sparsity on various data sets, including high-dimensional biological data. Our experimental results show that SLCE has a performance advantage over some state-of-the-art neural network-based feature selection techniques.
Feature Selection using Sparse Adaptive Bottleneck Centroid-Encoder
Jun 09, 2023Abstract:We introduce a novel nonlinear model, Sparse Adaptive Bottleneck Centroid-Encoder (SABCE), for determining the features that discriminate between two or more classes. The algorithm aims to extract discriminatory features in groups while reconstructing the class centroids in the ambient space and simultaneously use additional penalty terms in the bottleneck layer to decrease within-class scatter and increase the separation of different class centroids. The model has a sparsity-promoting layer (SPL) with a one-to-one connection to the input layer. Along with the primary objective, we minimize the $l_{2,1}$-norm of the sparse layer, which filters out unnecessary features from input data. During training, we update class centroids by taking the Hadamard product of the centroids and weights of the sparse layer, thus ignoring the irrelevant features from the target. Therefore the proposed method learns to reconstruct the critical components of class centroids rather than the whole centroids. The algorithm is applied to various real-world data sets, including high-dimensional biological, image, speech, and accelerometer sensor data. We compared our method to different state-of-the-art feature selection techniques, including supervised Concrete Autoencoders (SCAE), Feature Selection Networks (FsNet), Stochastic Gates (STG), and LassoNet. We empirically showed that SABCE features often produced better classification accuracy than other methods on the sequester test sets, setting new state-of-the-art results.
Yet Another Algorithm for Supervised Principal Component Analysis: Supervised Linear Centroid-Encoder
Jun 07, 2023



Abstract:We propose a new supervised dimensionality reduction technique called Supervised Linear Centroid-Encoder (SLCE), a linear counterpart of the nonlinear Centroid-Encoder (CE) \citep{ghosh2022supervised}. SLCE works by mapping the samples of a class to its class centroid using a linear transformation. The transformation is a projection that reconstructs a point such that its distance from the corresponding class centroid, i.e., centroid-reconstruction loss, is minimized in the ambient space. We derive a closed-form solution using an eigendecomposition of a symmetric matrix. We did a detailed analysis and presented some crucial mathematical properties of the proposed approach. %We also provide an iterative solution approach based solving the optimization problem using a descent method. We establish a connection between the eigenvalues and the centroid-reconstruction loss. In contrast to Principal Component Analysis (PCA) which reconstructs a sample in the ambient space, the transformation of SLCE uses the instances of a class to rebuild the corresponding class centroid. Therefore the proposed method can be considered a form of supervised PCA. Experimental results show the performance advantage of SLCE over other supervised methods.
Hamming Similarity and Graph Laplacians for Class Partitioning and Adversarial Image Detection
May 06, 2023



Abstract:Researchers typically investigate neural network representations by examining activation outputs for one or more layers of a network. Here, we investigate the potential for ReLU activation patterns (encoded as bit vectors) to aid in understanding and interpreting the behavior of neural networks. We utilize Representational Dissimilarity Matrices (RDMs) to investigate the coherence of data within the embedding spaces of a deep neural network. From each layer of a network, we extract and utilize bit vectors to construct similarity scores between images. From these similarity scores, we build a similarity matrix for a collection of images drawn from 2 classes. We then apply Fiedler partitioning to the associated Laplacian matrix to separate the classes. Our results indicate, through bit vector representations, that the network continues to refine class detectability with the last ReLU layer achieving better than 95\% separation accuracy. Additionally, we demonstrate that bit vectors aid in adversarial image detection, again achieving over 95\% accuracy in separating adversarial and non-adversarial images using a simple classifier.
Dual Graphs of Polyhedral Decompositions for the Detection of Adversarial Attacks
Dec 02, 2022Abstract:Previous work has shown that a neural network with the rectified linear unit (ReLU) activation function leads to a convex polyhedral decomposition of the input space. These decompositions can be represented by a dual graph with vertices corresponding to polyhedra and edges corresponding to polyhedra sharing a facet, which is a subgraph of a Hamming graph. This paper illustrates how one can utilize the dual graph to detect and analyze adversarial attacks in the context of digital images. When an image passes through a network containing ReLU nodes, the firing or non-firing at a node can be encoded as a bit ($1$ for ReLU activation, $0$ for ReLU non-activation). The sequence of all bit activations identifies the image with a bit vector, which identifies it with a polyhedron in the decomposition and, in turn, identifies it with a vertex in the dual graph. We identify ReLU bits that are discriminators between non-adversarial and adversarial images and examine how well collections of these discriminators can ensemble vote to build an adversarial image detector. Specifically, we examine the similarities and differences of ReLU bit vectors for adversarial images, and their non-adversarial counterparts, using a pre-trained ResNet-50 architecture. While this paper focuses on adversarial digital images, ResNet-50 architecture, and the ReLU activation function, our methods extend to other network architectures, activation functions, and types of datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge