Tomojit Ghosh
A Convex formulation for linear discriminant analysis
Mar 17, 2025Abstract:We present a supervised dimensionality reduction technique called Convex Linear Discriminant Analysis (ConvexLDA). The proposed model optimizes a multi-objective cost function by balancing two complementary terms. The first term pulls the samples of a class towards its centroid by minimizing a sample's distance from its class-centroid in low dimensional space. The second term pushes the classes far apart by maximizing their hyperellipsoid scattering volume via the logarithm of the determinant (\textit{log det}) of the outer product matrix formed by the low-dimensional class-centroids. Using the negative of the \textit{log det}, we pose the final cost as a minimization problem, which balances the two terms using a hyper-parameter $\lambda$. We demonstrate that the cost function is convex. Unlike Fisher LDA, the proposed method doesn't require to compute the inverse of a matrix, hence avoiding any ill-conditioned problem where data dimension is very high, e.g. RNA-seq data. ConvexLDA doesn't require pair-wise distance calculation, making it faster and more easily scalable. Moreover, the convex nature of the cost function ensures global optimality, enhancing the reliability of the learned embedding. Our experimental evaluation demonstrates that ConvexLDA outperforms several popular linear discriminant analysis (LDA)-based methods on a range of high-dimensional biological data, image data sets, etc.
A Multi-Domain Multi-Task Approach for Feature Selection from Bulk RNA Datasets
May 04, 2024Abstract:In this paper a multi-domain multi-task algorithm for feature selection in bulk RNAseq data is proposed. Two datasets are investigated arising from mouse host immune response to Salmonella infection. Data is collected from several strains of collaborative cross mice. Samples from the spleen and liver serve as the two domains. Several machine learning experiments are conducted and the small subset of discriminative across domains features have been extracted in each case. The algorithm proves viable and underlines the benefits of across domain feature selection by extracting new subset of discriminative features which couldn't be extracted only by one-domain approach.
Feature Selection using Sparse Adaptive Bottleneck Centroid-Encoder
Jun 09, 2023Abstract:We introduce a novel nonlinear model, Sparse Adaptive Bottleneck Centroid-Encoder (SABCE), for determining the features that discriminate between two or more classes. The algorithm aims to extract discriminatory features in groups while reconstructing the class centroids in the ambient space and simultaneously use additional penalty terms in the bottleneck layer to decrease within-class scatter and increase the separation of different class centroids. The model has a sparsity-promoting layer (SPL) with a one-to-one connection to the input layer. Along with the primary objective, we minimize the $l_{2,1}$-norm of the sparse layer, which filters out unnecessary features from input data. During training, we update class centroids by taking the Hadamard product of the centroids and weights of the sparse layer, thus ignoring the irrelevant features from the target. Therefore the proposed method learns to reconstruct the critical components of class centroids rather than the whole centroids. The algorithm is applied to various real-world data sets, including high-dimensional biological, image, speech, and accelerometer sensor data. We compared our method to different state-of-the-art feature selection techniques, including supervised Concrete Autoencoders (SCAE), Feature Selection Networks (FsNet), Stochastic Gates (STG), and LassoNet. We empirically showed that SABCE features often produced better classification accuracy than other methods on the sequester test sets, setting new state-of-the-art results.
Sparse Linear Centroid-Encoder: A Convex Method for Feature Selection
Jun 09, 2023Abstract:We present a novel feature selection technique, Sparse Linear Centroid-Encoder (SLCE). The algorithm uses a linear transformation to reconstruct a point as its class centroid and, at the same time, uses the $\ell_1$-norm penalty to filter out unnecessary features from the input data. The original formulation of the optimization problem is nonconvex, but we propose a two-step approach, where each step is convex. In the first step, we solve the linear Centroid-Encoder, a convex optimization problem over a matrix $A$. In the second step, we only search for a sparse solution over a diagonal matrix $B$ while keeping $A$ fixed. Unlike other linear methods, e.g., Sparse Support Vector Machines and Lasso, Sparse Linear Centroid-Encoder uses a single model for multi-class data. We present an in-depth empirical analysis of the proposed model and show that it promotes sparsity on various data sets, including high-dimensional biological data. Our experimental results show that SLCE has a performance advantage over some state-of-the-art neural network-based feature selection techniques.
Yet Another Algorithm for Supervised Principal Component Analysis: Supervised Linear Centroid-Encoder
Jun 07, 2023



Abstract:We propose a new supervised dimensionality reduction technique called Supervised Linear Centroid-Encoder (SLCE), a linear counterpart of the nonlinear Centroid-Encoder (CE) \citep{ghosh2022supervised}. SLCE works by mapping the samples of a class to its class centroid using a linear transformation. The transformation is a projection that reconstructs a point such that its distance from the corresponding class centroid, i.e., centroid-reconstruction loss, is minimized in the ambient space. We derive a closed-form solution using an eigendecomposition of a symmetric matrix. We did a detailed analysis and presented some crucial mathematical properties of the proposed approach. %We also provide an iterative solution approach based solving the optimization problem using a descent method. We establish a connection between the eigenvalues and the centroid-reconstruction loss. In contrast to Principal Component Analysis (PCA) which reconstructs a sample in the ambient space, the transformation of SLCE uses the instances of a class to rebuild the corresponding class centroid. Therefore the proposed method can be considered a form of supervised PCA. Experimental results show the performance advantage of SLCE over other supervised methods.
Sparse Centroid-Encoder: A Nonlinear Model for Feature Selection
Jan 30, 2022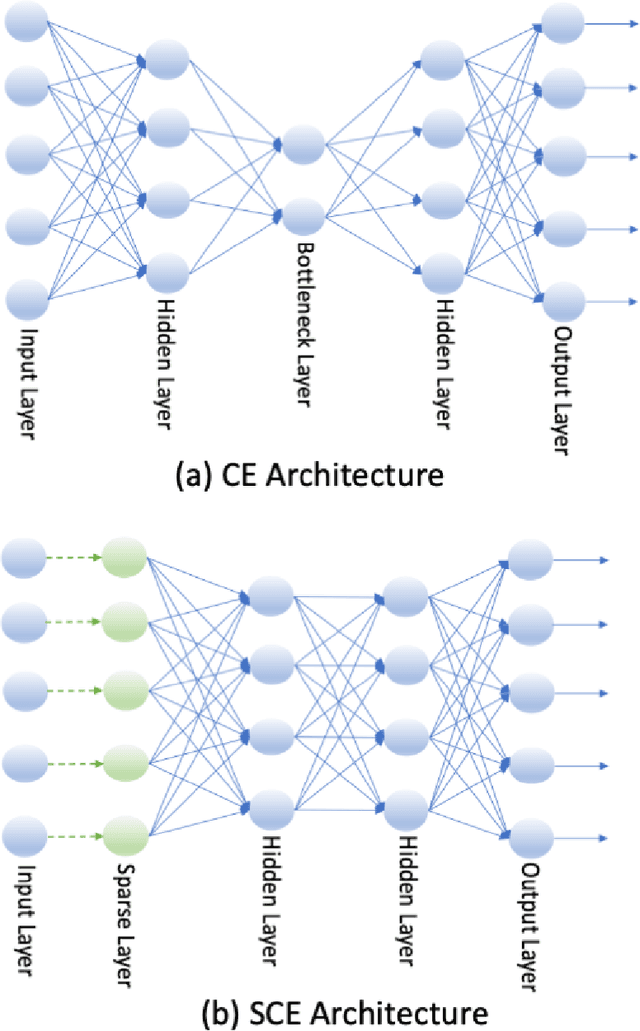


Abstract:We develop a sparse optimization problem for the determination of the total set of features that discriminate two or more classes. This is a sparse implementation of the centroid-encoder for nonlinear data reduction and visualization called Sparse Centroid-Encoder (SCE). We also provide a feature selection framework that first ranks each feature by its occurrence, and the optimal number of features is chosen using a validation set. The algorithm is applied to a wide variety of data sets including, single-cell biological data, high dimensional infectious disease data, hyperspectral data, image data, and speech data. We compared our method to various state-of-the-art feature selection techniques, including two neural network-based models (DFS, and LassoNet), Sparse SVM, and Random Forest. We empirically showed that SCE features produced better classification accuracy on the unseen test data, often with fewer features.
Supervised Dimensionality Reduction and Visualization using Centroid-encoder
Feb 28, 2020



Abstract:Visualizing high-dimensional data is an essential task in Data Science and Machine Learning. The Centroid-Encoder (CE) method is similar to the autoencoder but incorporates label information to keep objects of a class close together in the reduced visualization space. CE exploits nonlinearity and labels to encode high variance in low dimensions while capturing the global structure of the data. We present a detailed analysis of the method using a wide variety of data sets and compare it with other supervised dimension reduction techniques, including NCA, nonlinear NCA, t-distributed NCA, t-distributed MCML, supervised UMAP, supervised PCA, Colored Maximum Variance Unfolding, supervised Isomap, Parametric Embedding, supervised Neighbor Retrieval Visualizer, and Multiple Relational Embedding. We empirically show that centroid-encoder outperforms most of these techniques. We also show that when the data variance is spread across multiple modalities, centroid-encoder extracts a significant amount of information from the data in low dimensional space. This key feature establishes its value to use it as a tool for data visualization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge