Michael Habeck
Scaling Up Unbiased Search-based Symbolic Regression
Jun 24, 2025Abstract:In a regression task, a function is learned from labeled data to predict the labels at new data points. The goal is to achieve small prediction errors. In symbolic regression, the goal is more ambitious, namely, to learn an interpretable function that makes small prediction errors. This additional goal largely rules out the standard approach used in regression, that is, reducing the learning problem to learning parameters of an expansion of basis functions by optimization. Instead, symbolic regression methods search for a good solution in a space of symbolic expressions. To cope with the typically vast search space, most symbolic regression methods make implicit, or sometimes even explicit, assumptions about its structure. Here, we argue that the only obvious structure of the search space is that it contains small expressions, that is, expressions that can be decomposed into a few subexpressions. We show that systematically searching spaces of small expressions finds solutions that are more accurate and more robust against noise than those obtained by state-of-the-art symbolic regression methods. In particular, systematic search outperforms state-of-the-art symbolic regressors in terms of its ability to recover the true underlying symbolic expressions on established benchmark data sets.
Diffusion priors for Bayesian 3D reconstruction from incomplete measurements
Dec 19, 2024
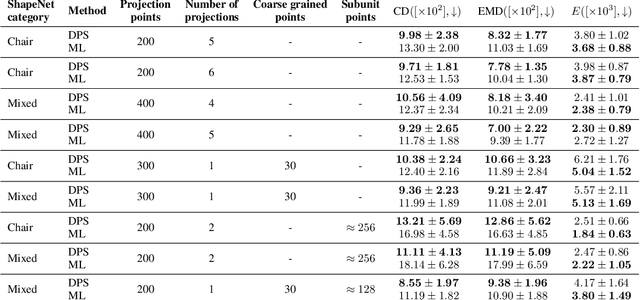

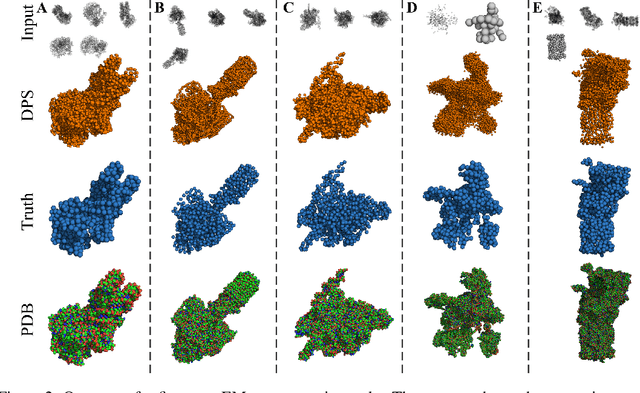
Abstract:Many inverse problems are ill-posed and need to be complemented by prior information that restricts the class of admissible models. Bayesian approaches encode this information as prior distributions that impose generic properties on the model such as sparsity, non-negativity or smoothness. However, in case of complex structured models such as images, graphs or three-dimensional (3D) objects,generic prior distributions tend to favor models that differ largely from those observed in the real world. Here we explore the use of diffusion models as priors that are combined with experimental data within a Bayesian framework. We use 3D point clouds to represent 3D objects such as household items or biomolecular complexes formed from proteins and nucleic acids. We train diffusion models that generate coarse-grained 3D structures at a medium resolution and integrate these with incomplete and noisy experimental data. To demonstrate the power of our approach, we focus on the reconstruction of biomolecular assemblies from cryo-electron microscopy (cryo-EM) images, which is an important inverse problem in structural biology. We find that posterior sampling with diffusion model priors allows for 3D reconstruction from very sparse, low-resolution and partial observations.
Bayesian multi-exposure image fusion for robust high dynamic range ptychography
Mar 19, 2024Abstract:Image information is restricted by the dynamic range of the detector, which can be addressed using multi-exposure image fusion (MEF). The conventional MEF approach employed in ptychography is often inadequate under conditions of low signal-to-noise ratio (SNR) or variations in illumination intensity. To address this, we developed a Bayesian approach for MEF based on a modified Poisson noise model that considers the background and saturation. Our method outperforms conventional MEF under challenging experimental conditions, as demonstrated by the synthetic and experimental data. Furthermore, this method is versatile and applicable to any imaging scheme requiring high dynamic range (HDR).
Parallel Affine Transformation Tuning of Markov Chain Monte Carlo
Jan 29, 2024Abstract:The performance of Markov chain Monte Carlo samplers strongly depends on the properties of the target distribution such as its covariance structure, the location of its probability mass and its tail behavior. We explore the use of bijective affine transformations of the sample space to improve the properties of the target distribution and thereby the performance of samplers running in the transformed space. In particular, we propose a flexible and user-friendly scheme for adaptively learning the affine transformation during sampling. Moreover, the combination of our scheme with Gibbsian polar slice sampling is shown to produce samples of high quality at comparatively low computational cost in several settings based on real-world data.
Gibbsian polar slice sampling
Feb 08, 2023Abstract:Polar slice sampling (Roberts & Rosenthal, 2002) is a Markov chain approach for approximate sampling of distributions that is difficult, if not impossible, to implement efficiently, but behaves provably well with respect to the dimension. By updating the directional and radial components of chain iterates separately, we obtain a family of samplers that mimic polar slice sampling, and yet can be implemented efficiently. Numerical experiments for a variety of settings indicate that our proposed algorithm outperforms the two most closely related approaches, elliptical slice sampling (Murray et al., 2010) and hit-and-run uniform slice sampling (MacKay, 2003). We prove the well-definedness and convergence of our methods under suitable assumptions on the target distribution.
Bayesian Evidence and Model Selection
Nov 23, 2015

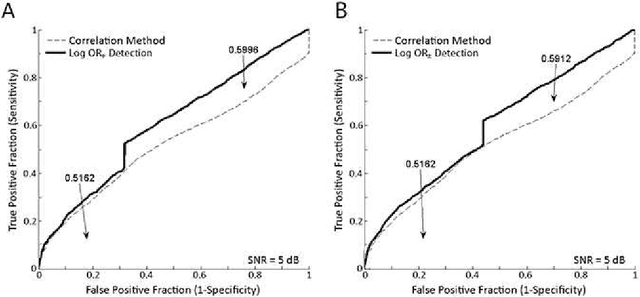

Abstract:In this paper we review the concepts of Bayesian evidence and Bayes factors, also known as log odds ratios, and their application to model selection. The theory is presented along with a discussion of analytic, approximate and numerical techniques. Specific attention is paid to the Laplace approximation, variational Bayes, importance sampling, thermodynamic integration, and nested sampling and its recent variants. Analogies to statistical physics, from which many of these techniques originate, are discussed in order to provide readers with deeper insights that may lead to new techniques. The utility of Bayesian model testing in the domain sciences is demonstrated by presenting four specific practical examples considered within the context of signal processing in the areas of signal detection, sensor characterization, scientific model selection and molecular force characterization.
* Arxiv version consists of 58 pages and 9 figures. Features theory, numerical methods and four applications
Adaptive nonparametric detection in cryo-electron microscopy
Nov 29, 2013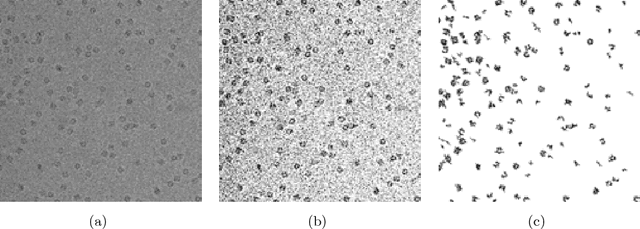
Abstract:Cryo-electron microscopy (cryo-EM) is an emerging experimental method to characterize the structure of large biomolecular assemblies. Single particle cryo-EM records 2D images (so-called micrographs) of projections of the three-dimensional particle, which need to be processed to obtain the three-dimensional reconstruction. A crucial step in the reconstruction process is particle picking which involves detection of particles in noisy 2D micrographs with low signal-to-noise ratios of typically 1:10 or even lower. Typically, each picture contains a large number of particles, and particles have unknown irregular and nonconvex shapes.
* Proceedings of the 58-th World Statistical Congress (2011)
Spatial statistics, image analysis and percolation theory
Oct 31, 2013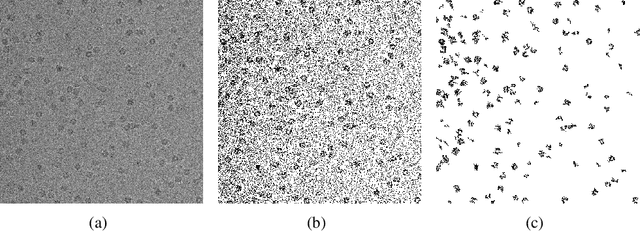
Abstract:We develop a novel method for detection of signals and reconstruction of images in the presence of random noise. The method uses results from percolation theory. We specifically address the problem of detection of multiple objects of unknown shapes in the case of nonparametric noise. The noise density is unknown and can be heavy-tailed. The objects of interest have unknown varying intensities. No boundary shape constraints are imposed on the objects, only a set of weak bulk conditions is required. We view the object detection problem as a multiple hypothesis testing for discrete statistical inverse problems. We present an algorithm that allows to detect greyscale objects of various shapes in noisy images. We prove results on consistency and algorithmic complexity of our procedures. Applications to cryo-electron microscopy are presented.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge