Mengdi Liu
Lost in Tokenization: Context as the Key to Unlocking Biomolecular Understanding in Scientific LLMs
Oct 27, 2025Abstract:Scientific Large Language Models (Sci-LLMs) have emerged as a promising frontier for accelerating biological discovery. However, these models face a fundamental challenge when processing raw biomolecular sequences: the tokenization dilemma. Whether treating sequences as a specialized language, risking the loss of functional motif information, or as a separate modality, introducing formidable alignment challenges, current strategies fundamentally limit their reasoning capacity. We challenge this sequence-centric paradigm by positing that a more effective strategy is to provide Sci-LLMs with high-level structured context derived from established bioinformatics tools, thereby bypassing the need to interpret low-level noisy sequence data directly. Through a systematic comparison of leading Sci-LLMs on biological reasoning tasks, we tested three input modes: sequence-only, context-only, and a combination of both. Our findings are striking: the context-only approach consistently and substantially outperforms all other modes. Even more revealing, the inclusion of the raw sequence alongside its high-level context consistently degrades performance, indicating that raw sequences act as informational noise, even for models with specialized tokenization schemes. These results suggest that the primary strength of existing Sci-LLMs lies not in their nascent ability to interpret biomolecular syntax from scratch, but in their profound capacity for reasoning over structured, human-readable knowledge. Therefore, we argue for reframing Sci-LLMs not as sequence decoders, but as powerful reasoning engines over expert knowledge. This work lays the foundation for a new class of hybrid scientific AI agents, repositioning the developmental focus from direct sequence interpretation towards high-level knowledge synthesis. The code is available at github.com/opendatalab-raise-dev/CoKE.
G2PDiffusion: Genotype-to-Phenotype Prediction with Diffusion Models
Feb 07, 2025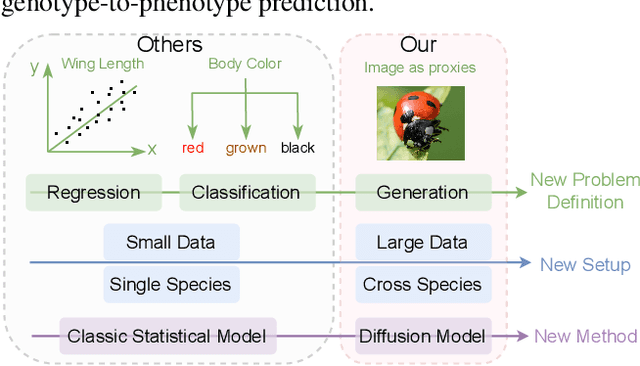
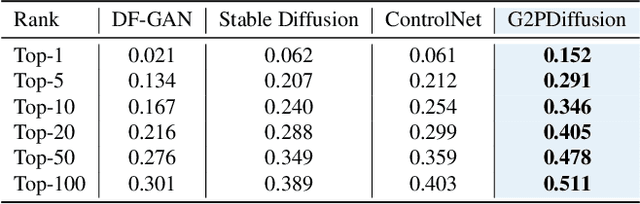
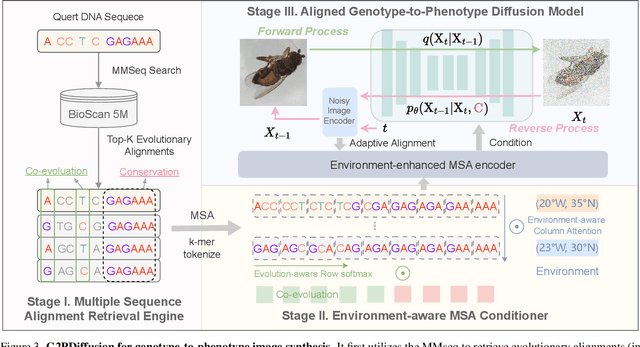
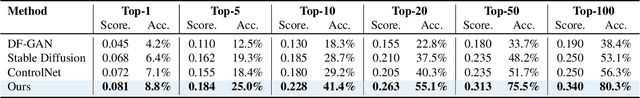
Abstract:Discovering the genotype-phenotype relationship is crucial for genetic engineering, which will facilitate advances in fields such as crop breeding, conservation biology, and personalized medicine. Current research usually focuses on single species and small datasets due to limitations in phenotypic data collection, especially for traits that require visual assessments or physical measurements. Deciphering complex and composite phenotypes, such as morphology, from genetic data at scale remains an open question. To break through traditional generic models that rely on simplified assumptions, this paper introduces G2PDiffusion, the first-of-its-kind diffusion model designed for genotype-to-phenotype generation across multiple species. Specifically, we use images to represent morphological phenotypes across species and redefine phenotype prediction as conditional image generation. To this end, this paper introduces an environment-enhanced DNA sequence conditioner and trains a stable diffusion model with a novel alignment method to improve genotype-to-phenotype consistency. Extensive experiments demonstrate that our approach enhances phenotype prediction accuracy across species, capturing subtle genetic variations that contribute to observable traits.
CP-DETR: Concept Prompt Guide DETR Toward Stronger Universal Object Detection
Dec 13, 2024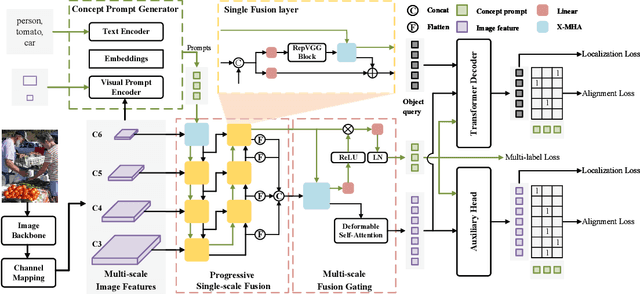
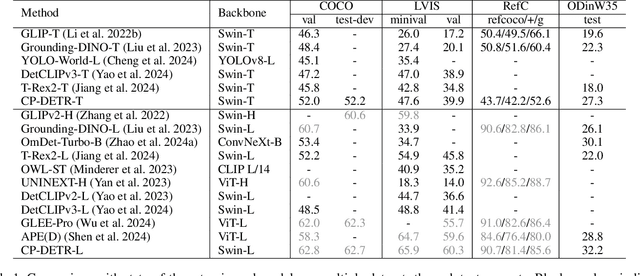
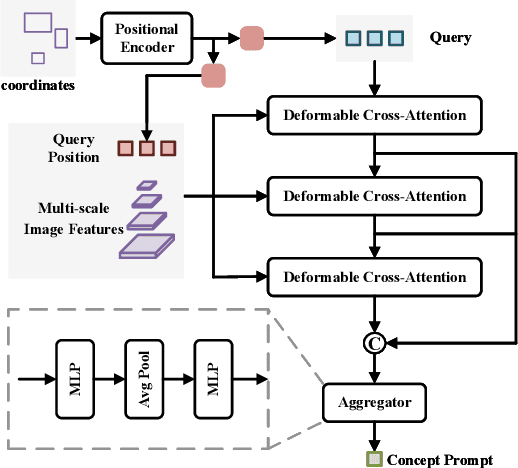
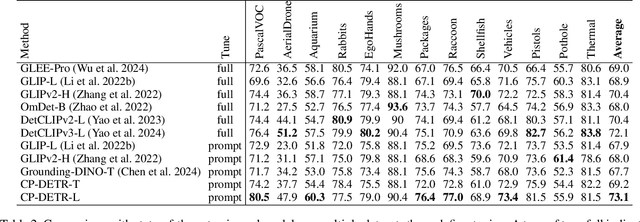
Abstract:Recent research on universal object detection aims to introduce language in a SoTA closed-set detector and then generalize the open-set concepts by constructing large-scale (text-region) datasets for training. However, these methods face two main challenges: (i) how to efficiently use the prior information in the prompts to genericise objects and (ii) how to reduce alignment bias in the downstream tasks, both leading to sub-optimal performance in some scenarios beyond pre-training. To address these challenges, we propose a strong universal detection foundation model called CP-DETR, which is competitive in almost all scenarios, with only one pre-training weight. Specifically, we design an efficient prompt visual hybrid encoder that enhances the information interaction between prompt and visual through scale-by-scale and multi-scale fusion modules. Then, the hybrid encoder is facilitated to fully utilize the prompted information by prompt multi-label loss and auxiliary detection head. In addition to text prompts, we have designed two practical concept prompt generation methods, visual prompt and optimized prompt, to extract abstract concepts through concrete visual examples and stably reduce alignment bias in downstream tasks. With these effective designs, CP-DETR demonstrates superior universal detection performance in a broad spectrum of scenarios. For example, our Swin-T backbone model achieves 47.6 zero-shot AP on LVIS, and the Swin-L backbone model achieves 32.2 zero-shot AP on ODinW35. Furthermore, our visual prompt generation method achieves 68.4 AP on COCO val by interactive detection, and the optimized prompt achieves 73.1 fully-shot AP on ODinW13.
Exploration of visual prompt in Grounded pre-trained open-set detection
Dec 14, 2023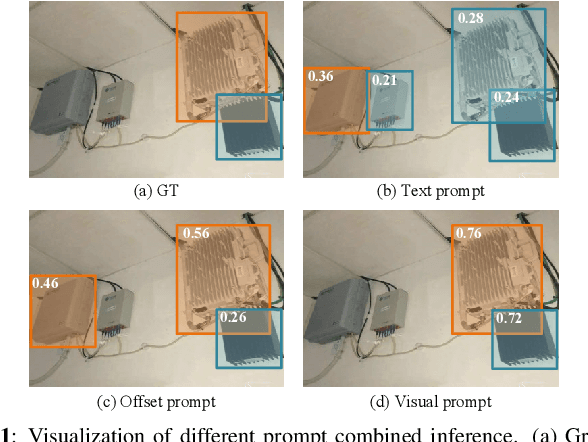
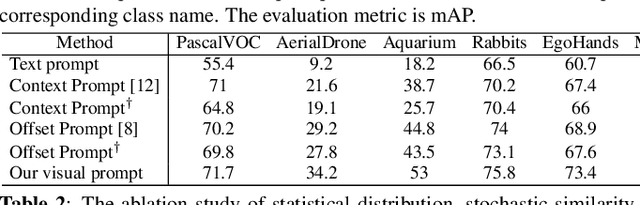
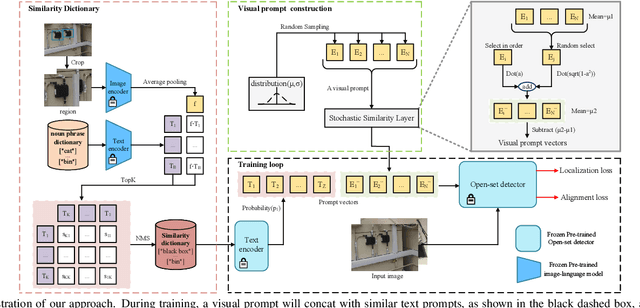
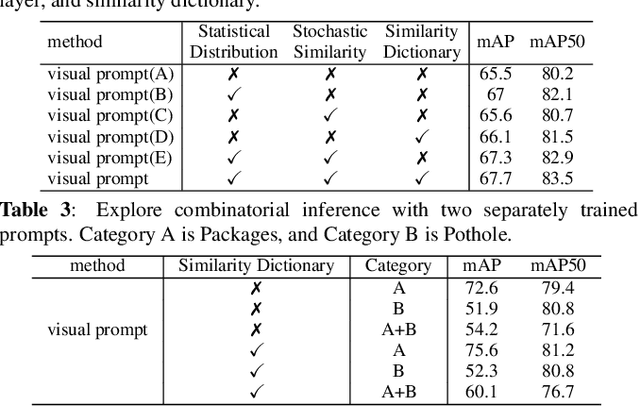
Abstract:Text prompts are crucial for generalizing pre-trained open-set object detection models to new categories. However, current methods for text prompts are limited as they require manual feedback when generalizing to new categories, which restricts their ability to model complex scenes, often leading to incorrect detection results. To address this limitation, we propose a novel visual prompt method that learns new category knowledge from a few labeled images, which generalizes the pre-trained detection model to the new category. To allow visual prompts to represent new categories adequately, we propose a statistical-based prompt construction module that is not limited by predefined vocabulary lengths, thus allowing more vectors to be used when representing categories. We further utilize the category dictionaries in the pre-training dataset to design task-specific similarity dictionaries, which make visual prompts more discriminative. We evaluate the method on the ODinW dataset and show that it outperforms existing prompt learning methods and performs more consistently in combinatorial inference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge