Masamichi Takahashi
Sketch-based Medical Image Retrieval
Mar 07, 2023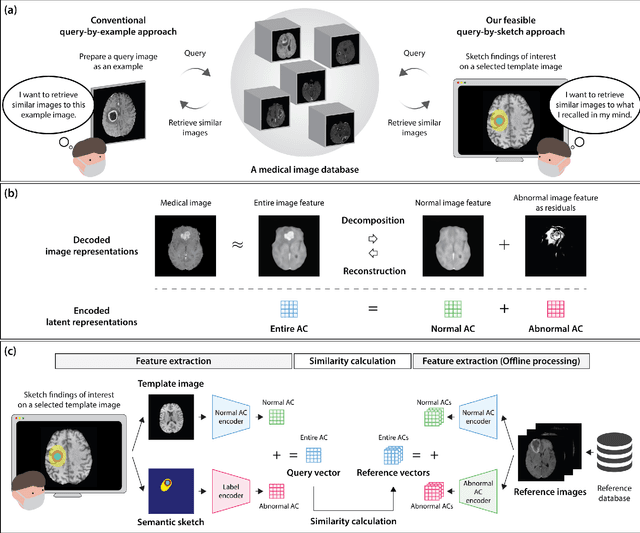
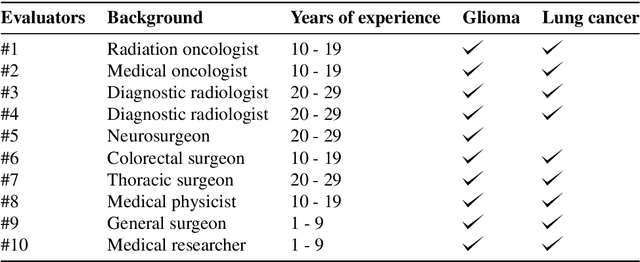
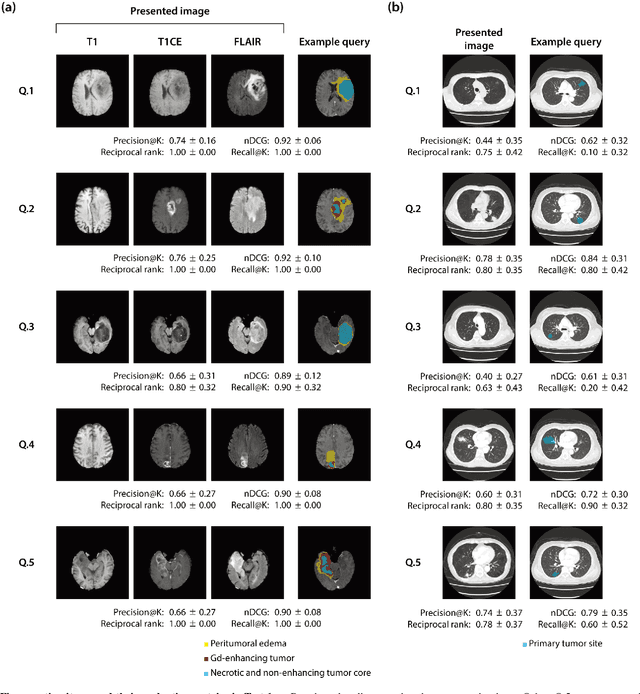
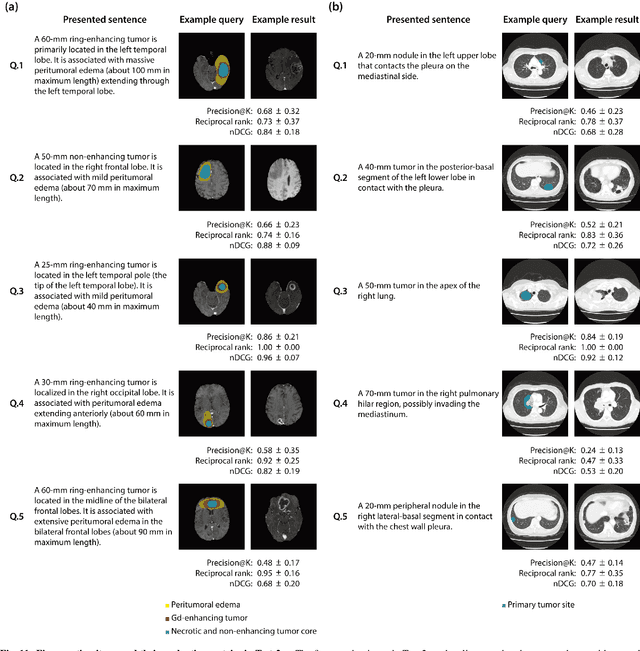
Abstract:The amount of medical images stored in hospitals is increasing faster than ever; however, utilizing the accumulated medical images has been limited. This is because existing content-based medical image retrieval (CBMIR) systems usually require example images to construct query vectors; nevertheless, example images cannot always be prepared. Besides, there can be images with rare characteristics that make it difficult to find similar example images, which we call isolated samples. Here, we introduce a novel sketch-based medical image retrieval (SBMIR) system that enables users to find images of interest without example images. The key idea lies in feature decomposition of medical images, whereby the entire feature of a medical image can be decomposed into and reconstructed from normal and abnormal features. By extending this idea, our SBMIR system provides an easy-to-use two-step graphical user interface: users first select a template image to specify a normal feature and then draw a semantic sketch of the disease on the template image to represent an abnormal feature. Subsequently, it integrates the two kinds of input to construct a query vector and retrieves reference images with the closest reference vectors. Using two datasets, ten healthcare professionals with various clinical backgrounds participated in the user test for evaluation. As a result, our SBMIR system enabled users to overcome previous challenges, including image retrieval based on fine-grained image characteristics, image retrieval without example images, and image retrieval for isolated samples. Our SBMIR system achieves flexible medical image retrieval on demand, thereby expanding the utility of medical image databases.
Decomposing Normal and Abnormal Features of Medical Images into Discrete Latent Codes for Content-Based Image Retrieval
Mar 23, 2021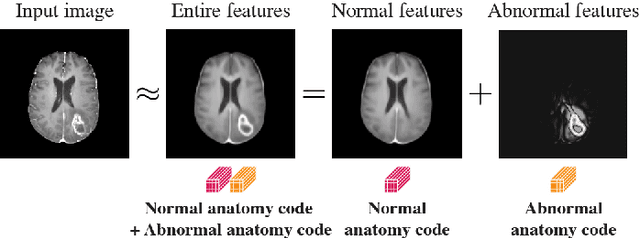
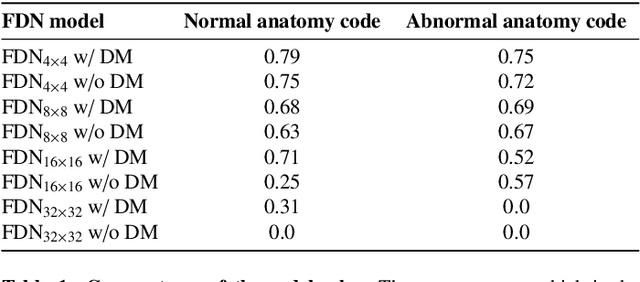
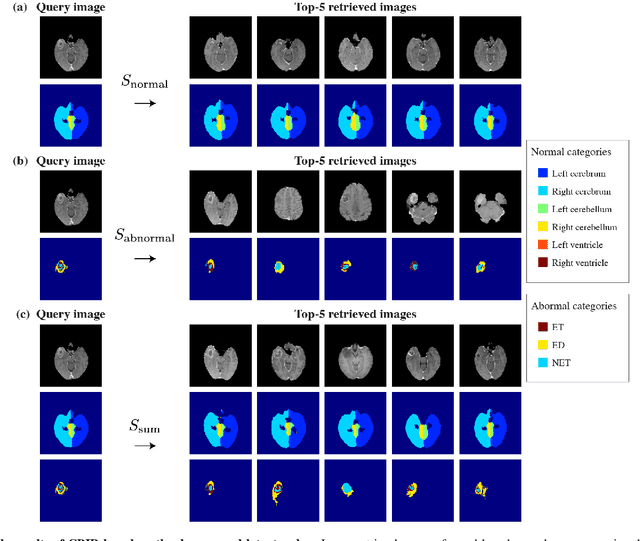
Abstract:In medical imaging, the characteristics purely derived from a disease should reflect the extent to which abnormal findings deviate from the normal features. Indeed, physicians often need corresponding images without abnormal findings of interest or, conversely, images that contain similar abnormal findings regardless of normal anatomical context. This is called comparative diagnostic reading of medical images, which is essential for a correct diagnosis. To support comparative diagnostic reading, content-based image retrieval (CBIR), which can selectively utilize normal and abnormal features in medical images as two separable semantic components, will be useful. Therefore, we propose a neural network architecture to decompose the semantic components of medical images into two latent codes: normal anatomy code and abnormal anatomy code. The normal anatomy code represents normal anatomies that should have existed if the sample is healthy, whereas the abnormal anatomy code attributes to abnormal changes that reflect deviation from the normal baseline. These latent codes are discretized through vector quantization to enable binary hashing, which can reduce the computational burden at the time of similarity search. By calculating the similarity based on either normal or abnormal anatomy codes or the combination of the two codes, our algorithm can retrieve images according to the selected semantic component from a dataset consisting of brain magnetic resonance images of gliomas. Our CBIR system qualitatively and quantitatively achieves remarkable results.
Unsupervised Brain Abnormality Detection Using High Fidelity Image Reconstruction Networks
Jun 02, 2020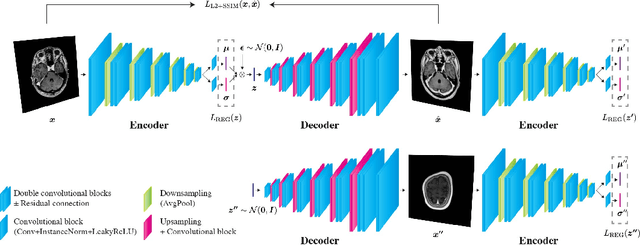
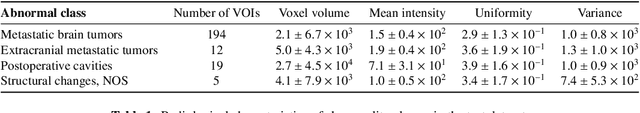
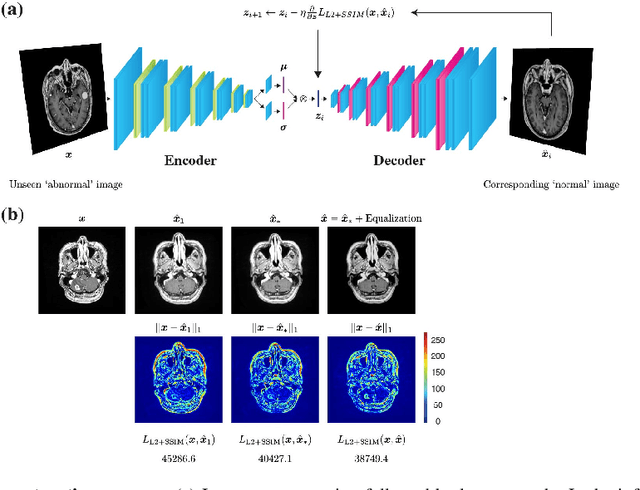
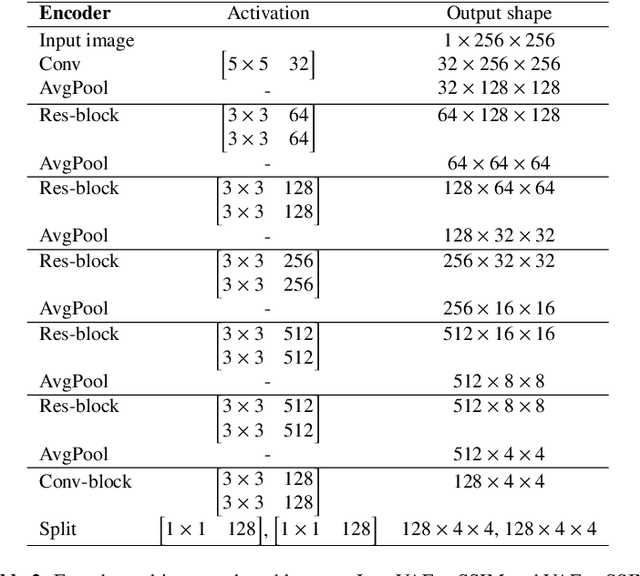
Abstract:Recent advances in deep learning have facilitated near-expert medical image analysis. Supervised learning is the mainstay of current approaches, though its success requires the use of large, fully labeled datasets. However, in real-world medical practice, previously unseen disease phenotypes are encountered that have not been defined a priori in finite-size datasets. Unsupervised learning, a hypothesis-free learning framework, may play a complementary role to supervised learning. Here, we demonstrate a novel framework for voxel-wise abnormality detection in brain magnetic resonance imaging (MRI), which exploits an image reconstruction network based on an introspective variational autoencoder trained with a structural similarity constraint. The proposed network learns a latent representation for "normal" anatomical variation using a series of images that do not include annotated abnormalities. After training, the network can map unseen query images to positions in the latent space, and latent variables sampled from those positions can be mapped back to the image space to yield normal-looking replicas of the input images. Finally, the network considers abnormality scores, which are designed to reflect differences at several image feature levels, in order to locate image regions that may contain abnormalities. The proposed method is evaluated on a comprehensively annotated dataset spanning clinically significant structural abnormalities of the brain parenchyma in a population having undergone radiotherapy for brain metastasis, demonstrating that it is particularly effective for contrast-enhanced lesions, i.e., metastatic brain tumors and extracranial metastatic tumors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge