Martin Hedlund
Segmenting Potentially Cancerous Areas in Prostate Biopsies using Semi-Automatically Annotated Data
Apr 15, 2019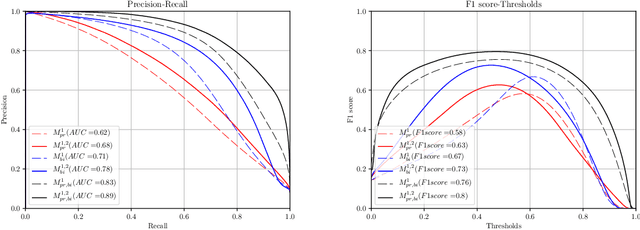
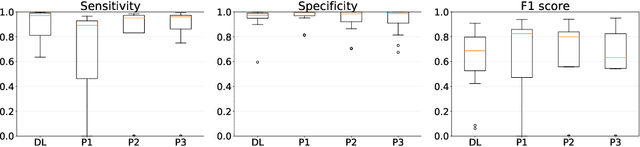
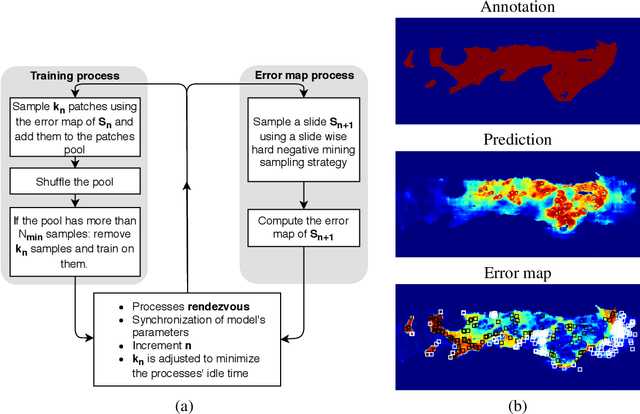

Abstract:Gleason grading specified in ISUP 2014 is the clinical standard in staging prostate cancer and the most important part of the treatment decision. However, the grading is subjective and suffers from high intra and inter-user variability. To improve the consistency and objectivity in the grading, we introduced glandular tissue WithOut Basal cells (WOB) as the ground truth. The presence of basal cells is the most accepted biomarker for benign glandular tissue and the absence of basal cells is a strong indicator of acinar prostatic adenocarcinoma, the most common form of prostate cancer. Glandular tissue can objectively be assessed as WOB or not WOB by using specific immunostaining for glandular tissue (Cytokeratin 8/18) and for basal cells (Cytokeratin 5/6 + p63). Even more, WOB allowed us to develop a semi-automated data generation pipeline to speed up the tremendously time consuming and expensive process of annotating whole slide images by pathologists. We generated 295 prostatectomy images exhaustively annotated with WOB. Then we used our Deep Learning Framework, which achieved the $2^{nd}$ best reported score in Camelyon17 Challenge, to train networks for segmenting WOB in needle biopsies. Evaluation of the model on 63 needle biopsies showed promising results which were improved further by finetuning the model on 118 biopsies annotated with WOB, achieving F1-score of 0.80 and Precision-Recall AUC of 0.89 at the pixel-level. Then we compared the performance of the model against 17 biopsies annotated independently by 3 pathologists using only H\&E staining. The comparison demonstrated that the model performed on a par with the pathologists. Finally, the model detected and accurately outlined existing WOB areas in two biopsies incorrectly annotated as totally WOB-free biopsies by three pathologists and in one biopsy by two pathologists.
Predicting breast tumor proliferation from whole-slide images: the TUPAC16 challenge
Jul 22, 2018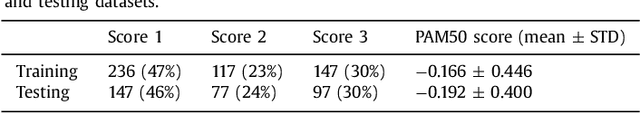
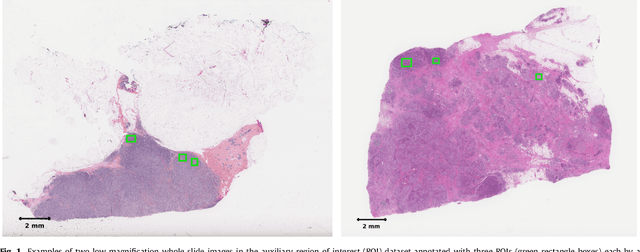
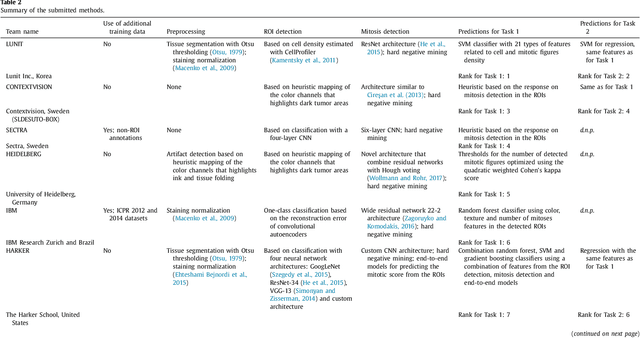
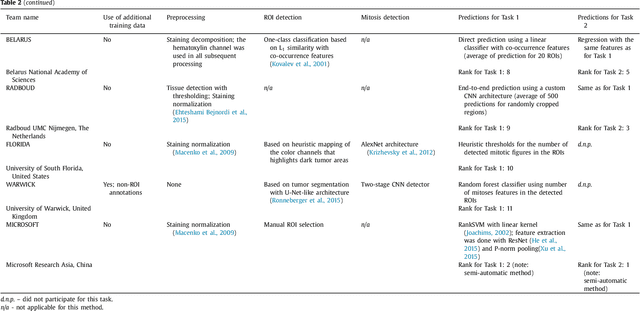
Abstract:Tumor proliferation is an important biomarker indicative of the prognosis of breast cancer patients. Assessment of tumor proliferation in a clinical setting is highly subjective and labor-intensive task. Previous efforts to automate tumor proliferation assessment by image analysis only focused on mitosis detection in predefined tumor regions. However, in a real-world scenario, automatic mitosis detection should be performed in whole-slide images (WSIs) and an automatic method should be able to produce a tumor proliferation score given a WSI as input. To address this, we organized the TUmor Proliferation Assessment Challenge 2016 (TUPAC16) on prediction of tumor proliferation scores from WSIs. The challenge dataset consisted of 500 training and 321 testing breast cancer histopathology WSIs. In order to ensure fair and independent evaluation, only the ground truth for the training dataset was provided to the challenge participants. The first task of the challenge was to predict mitotic scores, i.e., to reproduce the manual method of assessing tumor proliferation by a pathologist. The second task was to predict the gene expression based PAM50 proliferation scores from the WSI. The best performing automatic method for the first task achieved a quadratic-weighted Cohen's kappa score of $\kappa$ = 0.567, 95% CI [0.464, 0.671] between the predicted scores and the ground truth. For the second task, the predictions of the top method had a Spearman's correlation coefficient of r = 0.617, 95% CI [0.581 0.651] with the ground truth. This was the first study that investigated tumor proliferation assessment from WSIs. The achieved results are promising given the difficulty of the tasks and weakly-labelled nature of the ground truth. However, further research is needed to improve the practical utility of image analysis methods for this task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge