Nicolas Pinchaud
Weakly supervised training of pixel resolution segmentation models on whole slide images
May 30, 2019
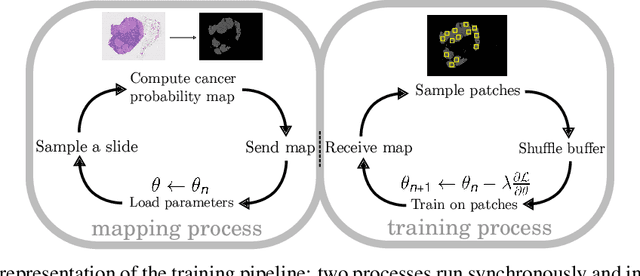
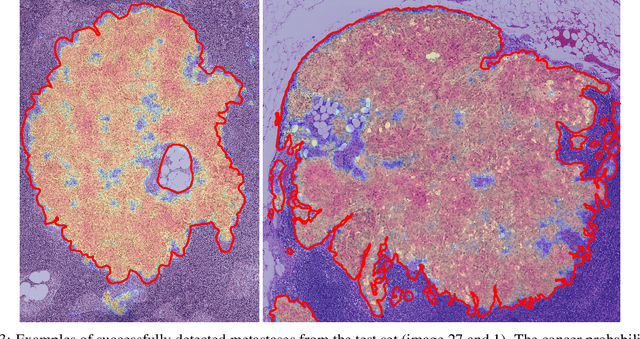
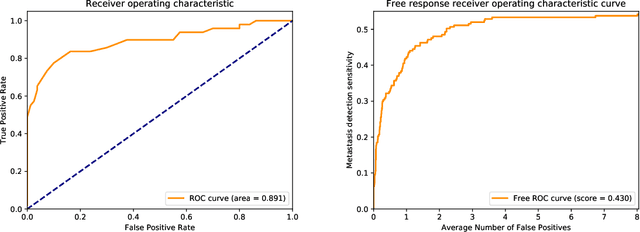
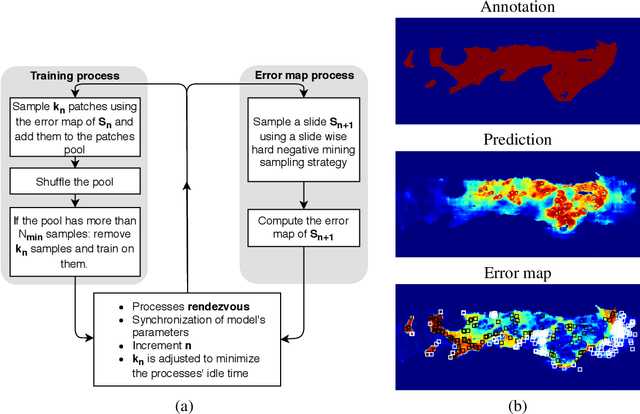
Abstract:We present a novel approach to train pixel resolution segmentation models on whole slide images in a weakly supervised setup. The model is trained to classify patches extracted from slides. This leads the training to be made under noisy labeled data. We solve the problem with two complementary strategies. First, the patches are sampled online using the model's knowledge by focusing on regions where the model's confidence is higher. Second, we propose an extension of the KL divergence that is robust to noisy labels. Our preliminary experiment on CAMELYON 16 data set show promising results. The model can successfully segment tumor areas with strong morphological consistency.
Unsupervised pre-training helps to conserve views from input distribution
May 30, 2019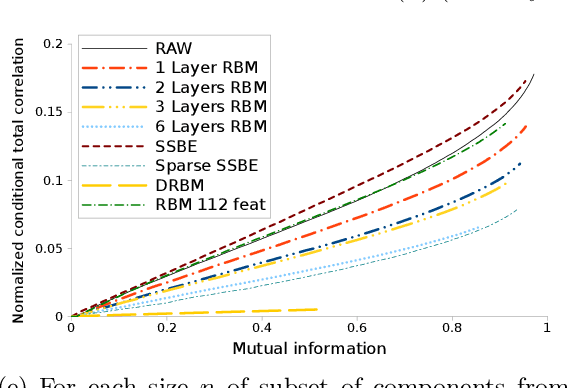
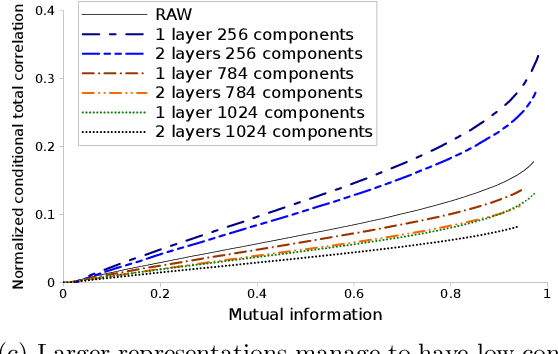
Abstract:We investigate the effects of the unsupervised pre-training method under the perspective of information theory. If the input distribution displays multiple views of the supervision, then unsupervised pre-training allows to learn hierarchical representation which communicates these views across layers, while disentangling the supervision. Disentanglement of supervision leads learned features to be independent conditionally to the label. In case of binary features, we show that conditional independence allows to extract label's information with a linear model and therefore helps to solve under-fitting. We suppose that representations displaying multiple views help to solve over-fitting because each view provides information that helps to reduce model's variance. We propose a practical method to measure both disentanglement of supervision and quantity of views within a binary representation. We show that unsupervised pre-training helps to conserve views from input distribution, whereas representations learned using supervised models disregard most of them.
Information theoretic learning of robust deep representations
May 30, 2019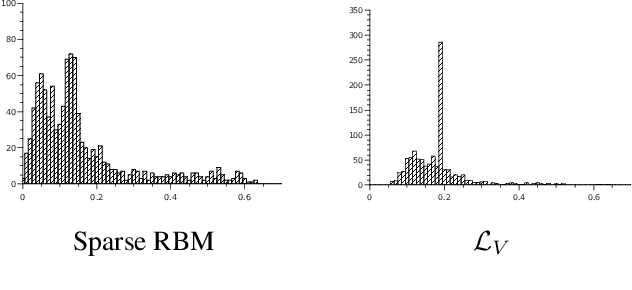

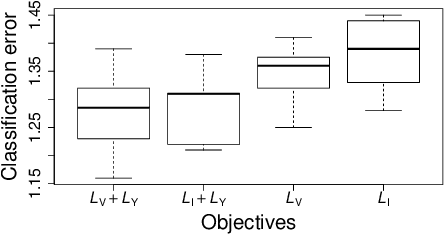
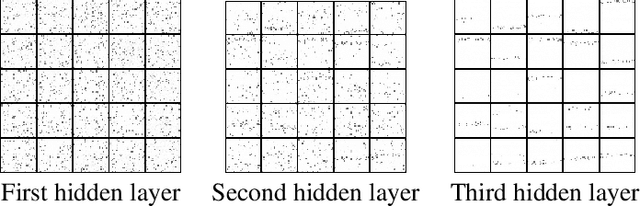
Abstract:We propose a novel objective function for learning robust deep representations of data based on information theory. Data is projected into a feature-vector space such that the mutual information of all subsets of features relative to the supervising signal is maximized. This objective function gives rise to robust representations by conserving available information relative to supervision in the face of noisy or unavailable features. Although the objective function is not directly tractable, we are able to derive a surrogate objective function. Minimizing this surrogate loss encourages features to be non-redundant and conditionally independent relative to the supervising signal. To evaluate the quality of obtained solutions, we have performed a set of preliminary experiments that show promising results.
Segmenting Potentially Cancerous Areas in Prostate Biopsies using Semi-Automatically Annotated Data
Apr 15, 2019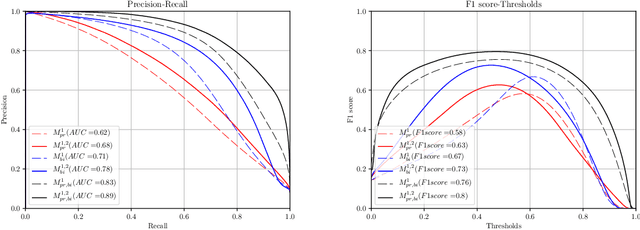
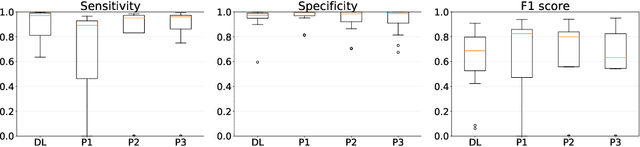

Abstract:Gleason grading specified in ISUP 2014 is the clinical standard in staging prostate cancer and the most important part of the treatment decision. However, the grading is subjective and suffers from high intra and inter-user variability. To improve the consistency and objectivity in the grading, we introduced glandular tissue WithOut Basal cells (WOB) as the ground truth. The presence of basal cells is the most accepted biomarker for benign glandular tissue and the absence of basal cells is a strong indicator of acinar prostatic adenocarcinoma, the most common form of prostate cancer. Glandular tissue can objectively be assessed as WOB or not WOB by using specific immunostaining for glandular tissue (Cytokeratin 8/18) and for basal cells (Cytokeratin 5/6 + p63). Even more, WOB allowed us to develop a semi-automated data generation pipeline to speed up the tremendously time consuming and expensive process of annotating whole slide images by pathologists. We generated 295 prostatectomy images exhaustively annotated with WOB. Then we used our Deep Learning Framework, which achieved the $2^{nd}$ best reported score in Camelyon17 Challenge, to train networks for segmenting WOB in needle biopsies. Evaluation of the model on 63 needle biopsies showed promising results which were improved further by finetuning the model on 118 biopsies annotated with WOB, achieving F1-score of 0.80 and Precision-Recall AUC of 0.89 at the pixel-level. Then we compared the performance of the model against 17 biopsies annotated independently by 3 pathologists using only H\&E staining. The comparison demonstrated that the model performed on a par with the pathologists. Finally, the model detected and accurately outlined existing WOB areas in two biopsies incorrectly annotated as totally WOB-free biopsies by three pathologists and in one biopsy by two pathologists.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge