Martin G. Skjæveland
PKG API: A Tool for Personal Knowledge Graph Management
Feb 12, 2024Abstract:Personal knowledge graphs (PKGs) offer individuals a way to store and consolidate their fragmented personal data in a central place, improving service personalization while maintaining full user control. Despite their potential, practical PKG implementations with user-friendly interfaces remain scarce. This work addresses this gap by proposing a complete solution to represent, manage, and interface with PKGs. Our approach includes (1) a user-facing PKG Client, enabling end-users to administer their personal data easily via natural language statements, and (2) a service-oriented PKG API. To tackle the complexity of representing these statements within a PKG, we present an RDF-based PKG vocabulary that supports this, along with properties for access rights and provenance.
An Ecosystem for Personal Knowledge Graphs: A Survey and Research Roadmap
Apr 19, 2023


Abstract:This paper presents an ecosystem for personal knowledge graphs (PKG), commonly defined as resources of structured information about entities related to an individual, their attributes, and the relations between them. PKGs are a key enabler of secure and sophisticated personal data management and personalized services. However, there are challenges that need to be addressed before PKGs can achieve widespread adoption. One of the fundamental challenges is the very definition of what constitutes a PKG, as there are multiple interpretations of the term. We propose our own definition of a PKG, emphasizing the aspects of (1) data ownership by a single individual and (2) the delivery of personalized services as the primary purpose. We further argue that a holistic view of PKGs is needed to unlock their full potential, and propose a unified framework for PKGs, where the PKG is a part of a larger ecosystem with clear interfaces towards data services and data sources. A comprehensive survey and synthesis of existing work is conducted, with a mapping of the surveyed work into the proposed unified ecosystem. Finally, we identify open challenges and research opportunities for the ecosystem as a whole, as well as for the specific aspects of PKGs, which include population, representation and management, and utilization.
A Survey of Syntactic Modelling Structures in Biomedical Ontologies
Jul 28, 2022
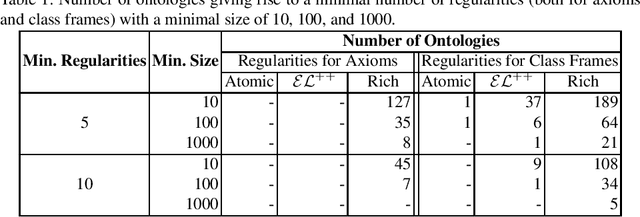
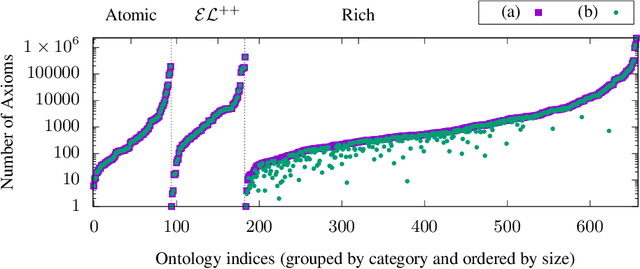
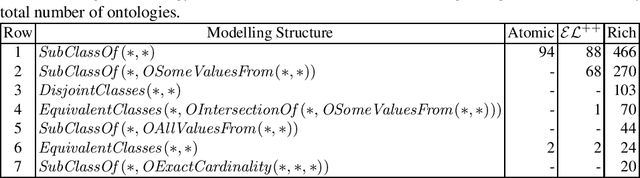
Abstract:Despite the large-scale uptake of semantic technologies in the biomedical domain, little is known about common modelling practices in published ontologies. OWL ontologies are often published only in the crude form of sets of axioms leaving the underlying design opaque. However, a principled and systematic ontology development life cycle is likely to be reflected in regularities of the ontology's emergent syntactic structure. To develop an understanding of this emergent structure, we propose to reverse-engineer ontologies taking a syntax-directed approach for identifying and analysing regularities for axioms and sets of axioms. We survey BioPortal in terms of syntactic modelling trends and common practices for OWL axioms and class frames. Our findings suggest that biomedical ontologies only share simple syntactic structures in which OWL constructors are not deeply nested or combined in a complex manner. While such simple structures often account for large proportions of axioms in a given ontology, many ontologies also contain non-trivial amounts of more complex syntactic structures that are not common across ontologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge