A Survey of Syntactic Modelling Structures in Biomedical Ontologies
Paper and Code
Jul 28, 2022
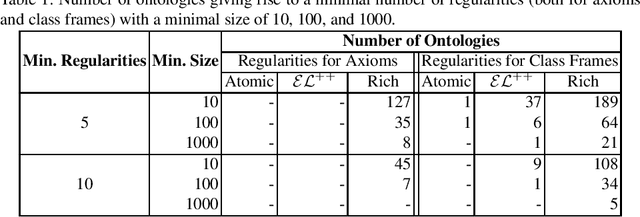
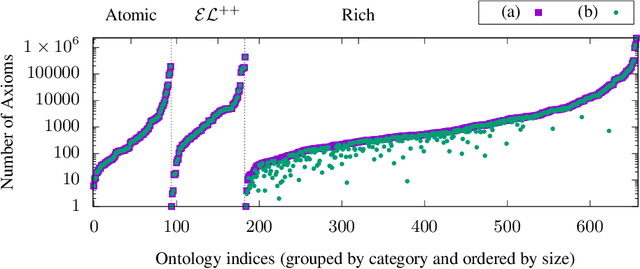
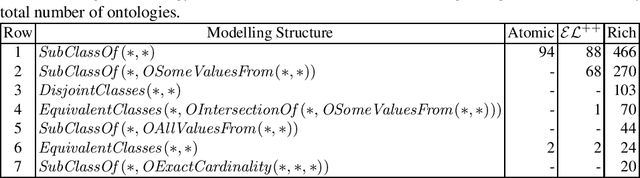
Despite the large-scale uptake of semantic technologies in the biomedical domain, little is known about common modelling practices in published ontologies. OWL ontologies are often published only in the crude form of sets of axioms leaving the underlying design opaque. However, a principled and systematic ontology development life cycle is likely to be reflected in regularities of the ontology's emergent syntactic structure. To develop an understanding of this emergent structure, we propose to reverse-engineer ontologies taking a syntax-directed approach for identifying and analysing regularities for axioms and sets of axioms. We survey BioPortal in terms of syntactic modelling trends and common practices for OWL axioms and class frames. Our findings suggest that biomedical ontologies only share simple syntactic structures in which OWL constructors are not deeply nested or combined in a complex manner. While such simple structures often account for large proportions of axioms in a given ontology, many ontologies also contain non-trivial amounts of more complex syntactic structures that are not common across ontologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge