Markus Stocker
Advancing Scientific Knowledge Retrieval and Reuse with a Novel Digital Library for Machine-Readable Knowledge
Nov 11, 2025Abstract:Digital libraries for research, such as the ACM Digital Library or Semantic Scholar, do not enable the machine-supported, efficient reuse of scientific knowledge (e.g., in synthesis research). This is because these libraries are based on document-centric models with narrative text knowledge expressions that require manual or semi-automated knowledge extraction, structuring, and organization. We present ORKG reborn, an emerging digital library that supports finding, accessing, and reusing accurate, fine-grained, and reproducible machine-readable expressions of scientific knowledge that relate scientific statements and their supporting evidence in terms of data and code. The rich expressions of scientific knowledge are published as reborn (born-reusable) articles and provide novel possibilities for scientific knowledge retrieval, for instance by statistical methods, software packages, variables, or data matching specific constraints. We describe the proposed system and demonstrate its practical viability and potential for information retrieval in contrast to state-of-the-art digital libraries and document-centric scholarly communication using several published articles in research fields ranging from computer science to soil science. Our work underscores the enormous potential of scientific knowledge databases and a viable approach to their construction.
Research Knowledge Graphs: the Shifting Paradigm of Scholarly Information Representation
Jun 08, 2025Abstract:Sharing and reusing research artifacts, such as datasets, publications, or methods is a fundamental part of scientific activity, where heterogeneity of resources and metadata and the common practice of capturing information in unstructured publications pose crucial challenges. Reproducibility of research and finding state-of-the-art methods or data have become increasingly challenging. In this context, the concept of Research Knowledge Graphs (RKGs) has emerged, aiming at providing an easy to use and machine-actionable representation of research artifacts and their relations. That is facilitated through the use of established principles for data representation, the consistent adoption of globally unique persistent identifiers and the reuse and linking of vocabularies and data. This paper provides the first conceptualisation of the RKG vision, a categorisation of in-use RKGs together with a description of RKG building blocks and principles. We also survey real-world RKG implementations differing with respect to scale, schema, data, used vocabulary, and reliability of the contained data. We also characterise different RKG construction methodologies and provide a forward-looking perspective on the diverse applications, opportunities, and challenges associated with the RKG vision.
SciCom Wiki: Fact-Checking and FAIR Knowledge Distribution for Scientific Videos and Podcasts
May 12, 2025Abstract:Democratic societies need accessible, reliable information. Videos and Podcasts have established themselves as the medium of choice for civic dissemination, but also as carriers of misinformation. The emerging Science Communication Knowledge Infrastructure (SciCom KI) curating non-textual media is still fragmented and not adequately equipped to scale against the content flood. Our work sets out to support the SciCom KI with a central, collaborative platform, the SciCom Wiki, to facilitate FAIR (findable, accessible, interoperable, reusable) media representation and the fact-checking of their content, particularly for videos and podcasts. Building an open-source service system centered around Wikibase, we survey requirements from 53 stakeholders, refine these in 11 interviews, and evaluate our prototype based on these requirements with another 14 participants. To address the most requested feature, fact-checking, we developed a neurosymbolic computational fact-checking approach, converting heterogenous media into knowledge graphs. This increases machine-readability and allows comparing statements against equally represented ground-truth. Our computational fact-checking tool was iteratively evaluated through 10 expert interviews, a public user survey with 43 participants verified the necessity and usability of our tool. Overall, our findings identified several needs to systematically support the SciCom KI. The SciCom Wiki, as a FAIR digital library complementing our neurosymbolic computational fact-checking framework, was found suitable to address the raised requirements. Further, we identified that the SciCom KI is severely underdeveloped regarding FAIR knowledge and related systems facilitating its collaborative creation and curation. Our system can provide a central knowledge node, yet a collaborative effort is required to scale against the imminent (mis-)information flood.
Computational Fact-Checking of Online Discourse: Scoring scientific accuracy in climate change related news articles
May 12, 2025Abstract:Democratic societies need reliable information. Misinformation in popular media such as news articles or videos threatens to impair civic discourse. Citizens are, unfortunately, not equipped to verify this content flood consumed daily at increasing rates. This work aims to semi-automatically quantify scientific accuracy of online media. By semantifying media of unknown veracity, their statements can be compared against equally processed trusted sources. We implemented a workflow using LLM-based statement extraction and knowledge graph analysis. Our neurosymbolic system was able to evidently streamline state-of-the-art veracity quantification. Evaluated via expert interviews and a user survey, the tool provides a beneficial veracity indication. This indicator, however, is unable to annotate public media at the required granularity and scale. Further work towards a FAIR (Findable, Accessible, Interoperable, Reusable) ground truth and complementary metrics are required to scientifically support civic discourse.
MORTY: Structured Summarization for Targeted Information Extraction from Scholarly Articles
Dec 11, 2022Abstract:Information extraction from scholarly articles is a challenging task due to the sizable document length and implicit information hidden in text, figures, and citations. Scholarly information extraction has various applications in exploration, archival, and curation services for digital libraries and knowledge management systems. We present MORTY, an information extraction technique that creates structured summaries of text from scholarly articles. Our approach condenses the article's full-text to property-value pairs as a segmented text snippet called structured summary. We also present a sizable scholarly dataset combining structured summaries retrieved from a scholarly knowledge graph and corresponding publicly available scientific articles, which we openly publish as a resource for the research community. Our results show that structured summarization is a suitable approach for targeted information extraction that complements other commonly used methods such as question answering and named entity recognition.
Plumber: A Modular Framework to Create Information Extraction Pipelines
Jun 03, 2022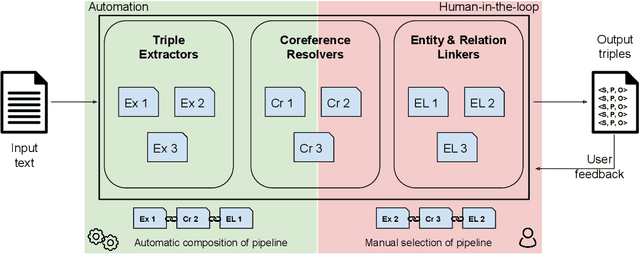
Abstract:Information Extraction (IE) tasks are commonly studied topics in various domains of research. Hence, the community continuously produces multiple techniques, solutions, and tools to perform such tasks. However, running those tools and integrating them within existing infrastructure requires time, expertise, and resources. One pertinent task here is triples extraction and linking, where structured triples are extracted from a text and aligned to an existing Knowledge Graph (KG). In this paper, we present PLUMBER, the first framework that allows users to manually and automatically create suitable IE pipelines from a community-created pool of tools to perform triple extraction and alignment on unstructured text. Our approach provides an interactive medium to alter the pipelines and perform IE tasks. A short video to show the working of the framework for different use-cases is available online under: https://www.youtube.com/watch?v=XC9rJNIUv8g
The Digitalization of Bioassays in the Open Research Knowledge Graph
Mar 28, 2022



Abstract:Background: Recent years are seeing a growing impetus in the semantification of scholarly knowledge at the fine-grained level of scientific entities in knowledge graphs. The Open Research Knowledge Graph (ORKG) https://www.orkg.org/ represents an important step in this direction, with thousands of scholarly contributions as structured, fine-grained, machine-readable data. There is a need, however, to engender change in traditional community practices of recording contributions as unstructured, non-machine-readable text. For this in turn, there is a strong need for AI tools designed for scientists that permit easy and accurate semantification of their scholarly contributions. We present one such tool, ORKG-assays. Implementation: ORKG-assays is a freely available AI micro-service in ORKG written in Python designed to assist scientists obtain semantified bioassays as a set of triples. It uses an AI-based clustering algorithm which on gold-standard evaluations over 900 bioassays with 5,514 unique property-value pairs for 103 predicates shows competitive performance. Results and Discussion: As a result, semantified assay collections can be surveyed on the ORKG platform via tabulation or chart-based visualizations of key property values of the chemicals and compounds offering smart knowledge access to biochemists and pharmaceutical researchers in the advancement of drug development.
Triple Classification for Scholarly Knowledge Graph Completion
Nov 23, 2021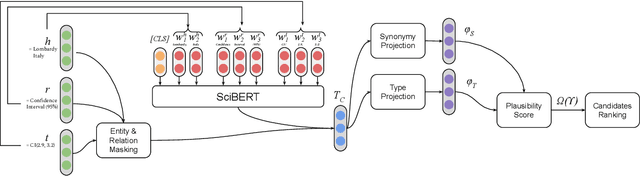

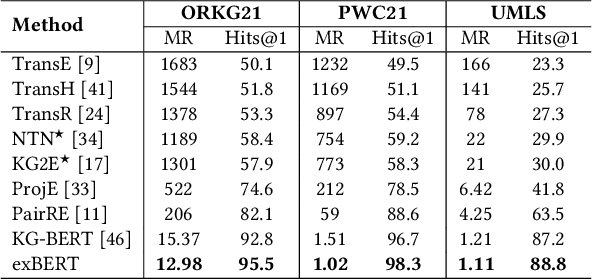
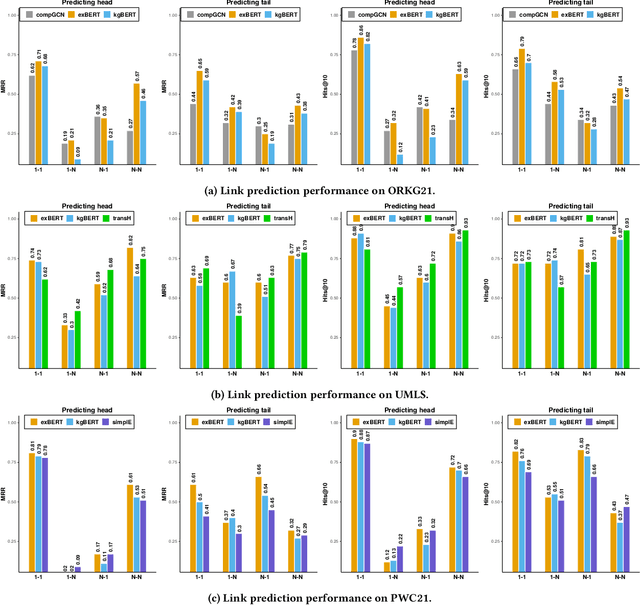
Abstract:Scholarly Knowledge Graphs (KGs) provide a rich source of structured information representing knowledge encoded in scientific publications. With the sheer volume of published scientific literature comprising a plethora of inhomogeneous entities and relations to describe scientific concepts, these KGs are inherently incomplete. We present exBERT, a method for leveraging pre-trained transformer language models to perform scholarly knowledge graph completion. We model triples of a knowledge graph as text and perform triple classification (i.e., belongs to KG or not). The evaluation shows that exBERT outperforms other baselines on three scholarly KG completion datasets in the tasks of triple classification, link prediction, and relation prediction. Furthermore, we present two scholarly datasets as resources for the research community, collected from public KGs and online resources.
Better Call the Plumber: Orchestrating Dynamic Information Extraction Pipelines
Feb 22, 2021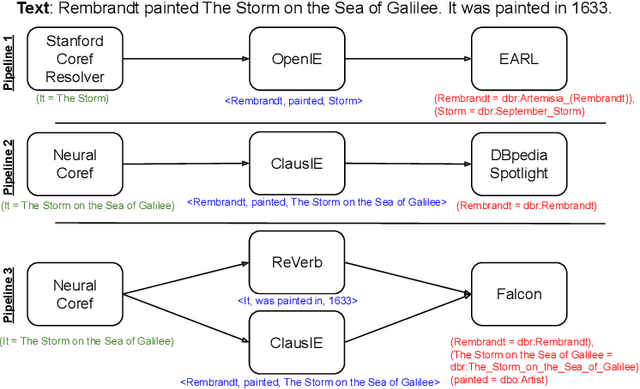
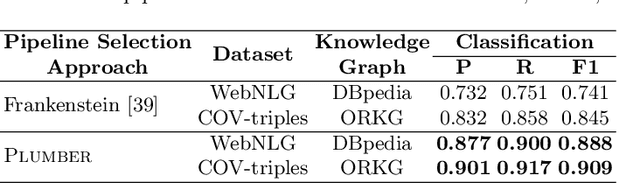
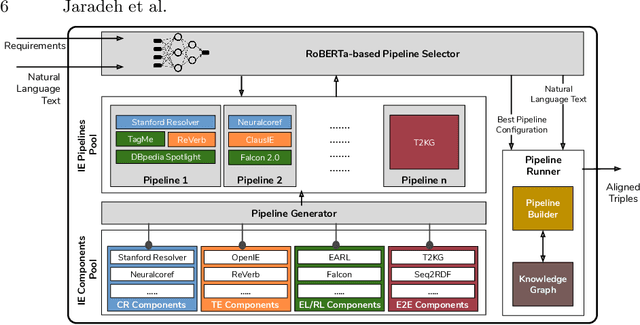
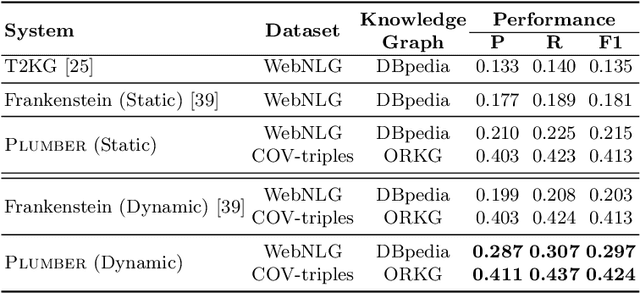
Abstract:In the last decade, a large number of Knowledge Graph (KG) information extraction approaches were proposed. Albeit effective, these efforts are disjoint, and their collective strengths and weaknesses in effective KG information extraction (IE) have not been studied in the literature. We propose Plumber, the first framework that brings together the research community's disjoint IE efforts. The Plumber architecture comprises 33 reusable components for various KG information extraction subtasks, such as coreference resolution, entity linking, and relation extraction. Using these components,Plumber dynamically generates suitable information extraction pipelines and offers overall 264 distinct pipelines.We study the optimization problem of choosing suitable pipelines based on input sentences. To do so, we train a transformer-based classification model that extracts contextual embeddings from the input and finds an appropriate pipeline. We study the efficacy of Plumber for extracting the KG triples using standard datasets over two KGs: DBpedia, and Open Research Knowledge Graph (ORKG). Our results demonstrate the effectiveness of Plumber in dynamically generating KG information extraction pipelines,outperforming all baselines agnostics of the underlying KG. Furthermore,we provide an analysis of collective failure cases, study the similarities and synergies among integrated components, and discuss their limitations.
Analysing the Requirements for an Open Research Knowledge Graph: Use Cases, Quality Requirements and Construction Strategies
Feb 11, 2021



Abstract:Current science communication has a number of drawbacks and bottlenecks which have been subject of discussion lately: Among others, the rising number of published articles makes it nearly impossible to get a full overview of the state of the art in a certain field, or reproducibility is hampered by fixed-length, document-based publications which normally cannot cover all details of a research work. Recently, several initiatives have proposed knowledge graphs (KG) for organising scientific information as a solution to many of the current issues. The focus of these proposals is, however, usually restricted to very specific use cases. In this paper, we aim to transcend this limited perspective and present a comprehensive analysis of requirements for an Open Research Knowledge Graph (ORKG) by (a) collecting and reviewing daily core tasks of a scientist, (b) establishing their consequential requirements for a KG-based system, (c) identifying overlaps and specificities, and their coverage in current solutions. As a result, we map necessary and desirable requirements for successful KG-based science communication, derive implications, and outline possible solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge