Arthur Brack
Citation Recommendation for Research Papers via Knowledge Graphs
Jun 10, 2021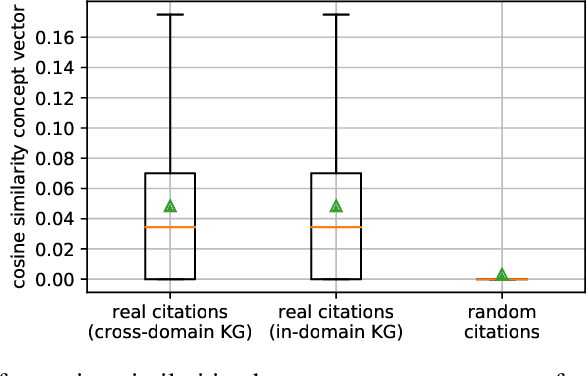

Abstract:Citation recommendation for research papers is a valuable task that can help researchers improve the quality of their work by suggesting relevant related work. Current approaches for this task rely primarily on the text of the papers and the citation network. In this paper, we propose to exploit an additional source of information, namely research knowledge graphs (KG) that interlink research papers based on mentioned scientific concepts. Our experimental results demonstrate that the combination of information from research KGs with existing state-of-the-art approaches is beneficial. Experimental results are presented for the STM-KG (STM: Science, Technology, Medicine), which is an automatically populated knowledge graph based on the scientific concepts extracted from papers of ten domains. The proposed approach outperforms the state of the art with a mean average precision of 20.6% (+0.8) for the top-50 retrieved results.
Analysing the Requirements for an Open Research Knowledge Graph: Use Cases, Quality Requirements and Construction Strategies
Feb 11, 2021



Abstract:Current science communication has a number of drawbacks and bottlenecks which have been subject of discussion lately: Among others, the rising number of published articles makes it nearly impossible to get a full overview of the state of the art in a certain field, or reproducibility is hampered by fixed-length, document-based publications which normally cannot cover all details of a research work. Recently, several initiatives have proposed knowledge graphs (KG) for organising scientific information as a solution to many of the current issues. The focus of these proposals is, however, usually restricted to very specific use cases. In this paper, we aim to transcend this limited perspective and present a comprehensive analysis of requirements for an Open Research Knowledge Graph (ORKG) by (a) collecting and reviewing daily core tasks of a scientist, (b) establishing their consequential requirements for a KG-based system, (c) identifying overlaps and specificities, and their coverage in current solutions. As a result, we map necessary and desirable requirements for successful KG-based science communication, derive implications, and outline possible solutions.
Sequential Sentence Classification in Research Papers using Cross-Domain Multi-Task Learning
Feb 11, 2021



Abstract:The task of sequential sentence classification enables the semantic structuring of research papers. This can enhance academic search engines to support researchers in finding and exploring research literature more effectively. However, previous work has not investigated the potential of transfer learning with datasets from different scientific domains for this task yet. We propose a uniform deep learning architecture and multi-task learning to improve sequential sentence classification in scientific texts across domains by exploiting training data from multiple domains. Our contributions can be summarised as follows: (1) We tailor two common transfer learning methods, sequential transfer learning and multi-task learning, and evaluate their performance for sequential sentence classification; (2) The presented multi-task model is able to recognise semantically related classes from different datasets and thus supports manual comparison and assessment of different annotation schemes; (3) The unified approach is capable of handling datasets that contain either only abstracts or full papers without further feature engineering. We demonstrate that models, which are trained on datasets from different scientific domains, benefit from one another when using the proposed multi-task learning architecture. Our approach outperforms the state of the art on three benchmark datasets.
Coreference Resolution in Research Papers from Multiple Domains
Jan 04, 2021



Abstract:Coreference resolution is essential for automatic text understanding to facilitate high-level information retrieval tasks such as text summarisation or question answering. Previous work indicates that the performance of state-of-the-art approaches (e.g. based on BERT) noticeably declines when applied to scientific papers. In this paper, we investigate the task of coreference resolution in research papers and subsequent knowledge graph population. We present the following contributions: (1) We annotate a corpus for coreference resolution that comprises 10 different scientific disciplines from Science, Technology, and Medicine (STM); (2) We propose transfer learning for automatic coreference resolution in research papers; (3) We analyse the impact of coreference resolution on knowledge graph (KG) population; (4) We release a research KG that is automatically populated from 55,485 papers in 10 STM domains. Comprehensive experiments show the usefulness of the proposed approach. Our transfer learning approach considerably outperforms state-of-the-art baselines on our corpus with an F1 score of 61.4 (+11.0), while the evaluation against a gold standard KG shows that coreference resolution improves the quality of the populated KG significantly with an F1 score of 63.5 (+21.8).
The STEM-ECR Dataset: Grounding Scientific Entity References in STEM Scholarly Content to Authoritative Encyclopedic and Lexicographic Sources
Mar 06, 2020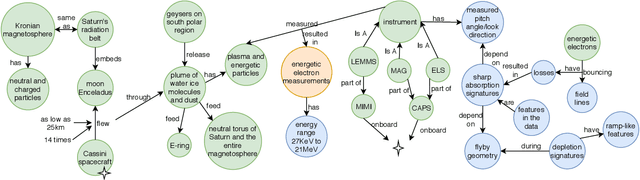
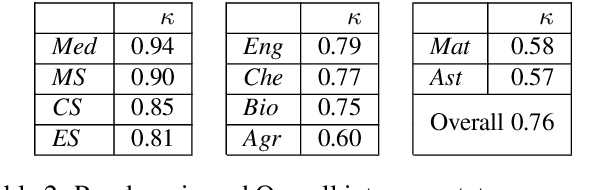
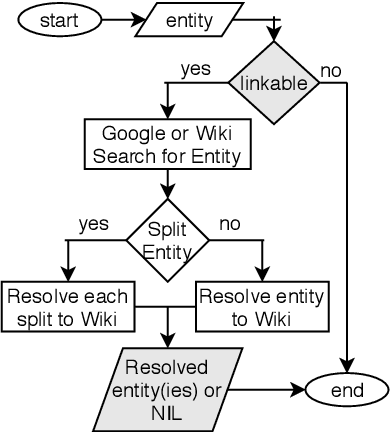
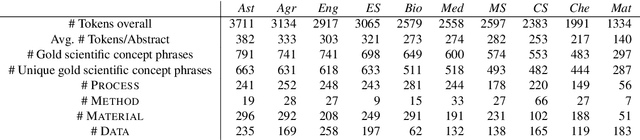
Abstract:We introduce the STEM (Science, Technology, Engineering, and Medicine) Dataset for Scientific Entity Extraction, Classification, and Resolution, version 1.0 (STEM-ECR v1.0). The STEM-ECR v1.0 dataset has been developed to provide a benchmark for the evaluation of scientific entity extraction, classification, and resolution tasks in a domain-independent fashion. It comprises abstracts in 10 STEM disciplines that were found to be the most prolific ones on a major publishing platform. We describe the creation of such a multidisciplinary corpus and highlight the obtained findings in terms of the following features: 1) a generic conceptual formalism for scientific entities in a multidisciplinary scientific context; 2) the feasibility of the domain-independent human annotation of scientific entities under such a generic formalism; 3) a performance benchmark obtainable for automatic extraction of multidisciplinary scientific entities using BERT-based neural models; 4) a delineated 3-step entity resolution procedure for human annotation of the scientific entities via encyclopedic entity linking and lexicographic word sense disambiguation; and 5) human evaluations of Babelfy returned encyclopedic links and lexicographic senses for our entities. Our findings cumulatively indicate that human annotation and automatic learning of multidisciplinary scientific concepts as well as their semantic disambiguation in a wide-ranging setting as STEM is reasonable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge