Mark A. van de Wiel
Hypothesis-free discovery from epidemiological data by automatic detection and local inference for tree-based nonlinearities and interactions
May 01, 2025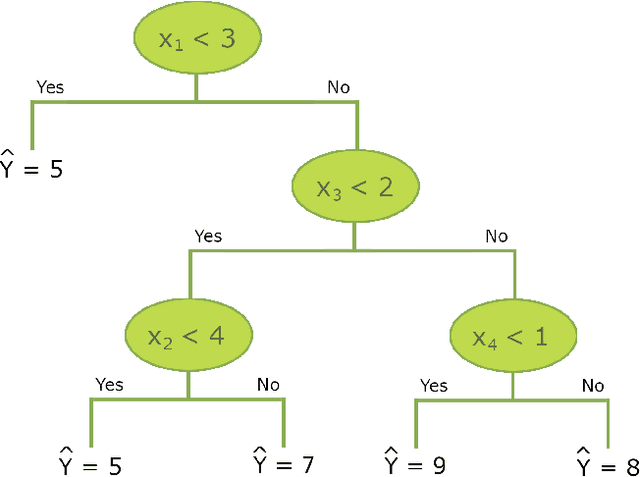

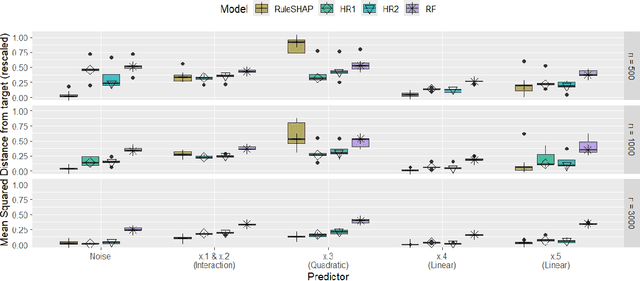
Abstract:In epidemiological settings, Machine Learning (ML) is gaining popularity for hypothesis-free discovery of risk (or protective) factors. Although ML is strong at discovering non-linearities and interactions, this power is currently compromised by a lack of reliable inference. Although local measures of feature effect can be combined with tree ensembles, uncertainty quantifications for these measures remain only partially available and oftentimes unsatisfactory. We propose RuleSHAP, a framework for using rule-based, hypothesis-free discovery that combines sparse Bayesian regression, tree ensembles and Shapley values in a one-step procedure that both detects and tests complex patterns at the individual level. To ease computation, we derive a formula that computes marginal Shapley values more efficiently for our setting. We demonstrate the validity of our framework on simulated data. To illustrate, we apply our machinery to data from an epidemiological cohort to detect and infer several effects for high cholesterol and blood pressure, such as nonlinear interaction effects between features like age, sex, ethnicity, BMI and glucose level.
Refining CART Models for Covariate Shift with Importance Weight
Oct 28, 2024Abstract:Machine learning models often face challenges in medical applications due to covariate shifts, where discrepancies between training and target data distributions can decrease predictive accuracy. This paper introduces an adaptation of Classification and Regression Trees (CART) that incorporates importance weighting to address these distributional differences effectively. By assigning greater weight to training samples that closely represent the target distribution, our approach modifies the CART model to improve performance in the presence of covariate shift. We evaluate the effectiveness of this method through simulation studies and apply it to real-world medical data, showing significant improvements in predictive accuracy. The results indicate that this weighted CART approach can be valuable in medical and other fields where covariate shift poses challenges, enabling more reliable predictions across diverse data distributions.
Guiding adaptive shrinkage by co-data to improve regression-based prediction and feature selection
May 08, 2024Abstract:The high dimensional nature of genomics data complicates feature selection, in particular in low sample size studies - not uncommon in clinical prediction settings. It is widely recognized that complementary data on the features, `co-data', may improve results. Examples are prior feature groups or p-values from a related study. Such co-data are ubiquitous in genomics settings due to the availability of public repositories. Yet, the uptake of learning methods that structurally use such co-data is limited. We review guided adaptive shrinkage methods: a class of regression-based learners that use co-data to adapt the shrinkage parameters, crucial for the performance of those learners. We discuss technical aspects, but also the applicability in terms of types of co-data that can be handled. This class of methods is contrasted with several others. In particular, group-adaptive shrinkage is compared with the better-known sparse group-lasso by evaluating feature selection. Finally, we demonstrate the versatility of the guided shrinkage methodology by showing how to `do-it-yourself': we integrate implementations of a co-data learner and the spike-and-slab prior for the purpose of improving feature selection in genetics studies.
Co-data Learning for Bayesian Additive Regression Trees
Nov 16, 2023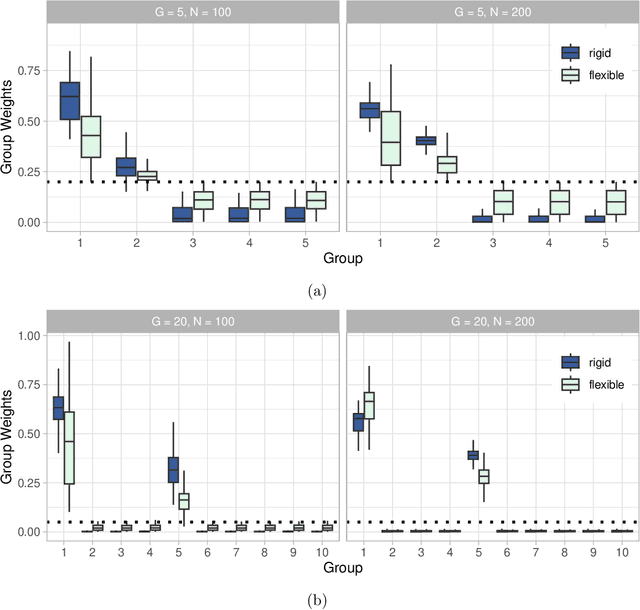
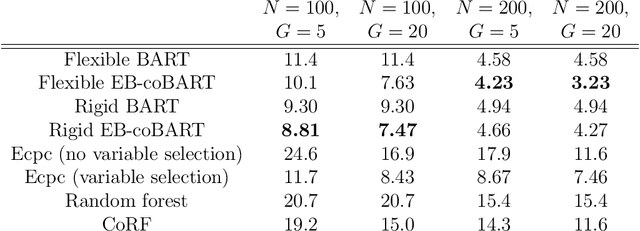
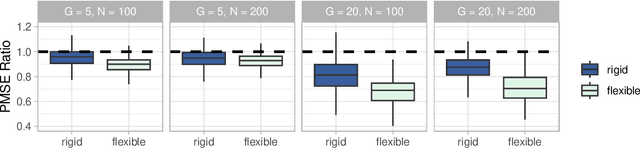
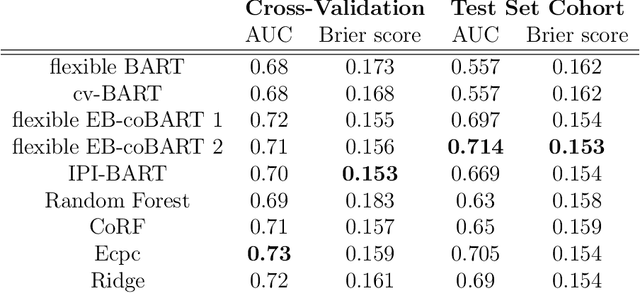
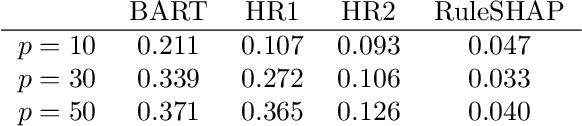
Abstract:Medical prediction applications often need to deal with small sample sizes compared to the number of covariates. Such data pose problems for prediction and variable selection, especially when the covariate-response relationship is complicated. To address these challenges, we propose to incorporate co-data, i.e. external information on the covariates, into Bayesian additive regression trees (BART), a sum-of-trees prediction model that utilizes priors on the tree parameters to prevent overfitting. To incorporate co-data, an empirical Bayes (EB) framework is developed that estimates, assisted by a co-data model, prior covariate weights in the BART model. The proposed method can handle multiple types of co-data simultaneously. Furthermore, the proposed EB framework enables the estimation of the other hyperparameters of BART as well, rendering an appealing alternative to cross-validation. We show that the method finds relevant covariates and that it improves prediction compared to default BART in simulations. If the covariate-response relationship is nonlinear, the method benefits from the flexibility of BART to outperform regression-based co-data learners. Finally, the use of co-data enhances prediction in an application to diffuse large B-cell lymphoma prognosis based on clinical covariates, gene mutations, DNA translocations, and DNA copy number data. Keywords: Bayesian additive regression trees; Empirical Bayes; Co-data; High-dimensional data; Omics; Prediction
Linked shrinkage to improve estimation of interaction effects in regression models
Sep 25, 2023Abstract:We address a classical problem in statistics: adding two-way interaction terms to a regression model. As the covariate dimension increases quadratically, we develop an estimator that adapts well to this increase, while providing accurate estimates and appropriate inference. Existing strategies overcome the dimensionality problem by only allowing interactions between relevant main effects. Building on this philosophy, we implement a softer link between the two types of effects using a local shrinkage model. We empirically show that borrowing strength between the amount of shrinkage for main effects and their interactions can strongly improve estimation of the regression coefficients. Moreover, we evaluate the potential of the model for inference, which is notoriously hard for selection strategies. Large-scale cohort data are used to provide realistic illustrations and evaluations. Comparisons with other methods are provided. The evaluation of variable importance is not trivial in regression models with many interaction terms. Therefore, we derive a new analytical formula for the Shapley value, which enables rapid assessment of individual-specific variable importance scores and their uncertainties. Finally, while not targeting for prediction, we do show that our models can be very competitive to a more advanced machine learner, like random forest, even for fairly large sample sizes. The implementation of our method in RStan is fairly straightforward, allowing for adjustments to specific needs.
Penalised regression with multiple sources of prior effects
Dec 16, 2022Abstract:In many high-dimensional prediction or classification tasks, complementary data on the features are available, e.g. prior biological knowledge on (epi)genetic markers. Here we consider tasks with numerical prior information that provide an insight into the importance (weight) and the direction (sign) of the feature effects, e.g. regression coefficients from previous studies. We propose an approach for integrating multiple sources of such prior information into penalised regression. If suitable co-data are available, this improves the predictive performance, as shown by simulation and application. The proposed method is implemented in the R package `transreg' (https://github.com/lcsb-bds/transreg).
Estimation of Predictive Performance in High-Dimensional Data Settings using Learning Curves
Jun 08, 2022



Abstract:In high-dimensional prediction settings, it remains challenging to reliably estimate the test performance. To address this challenge, a novel performance estimation framework is presented. This framework, called Learn2Evaluate, is based on learning curves by fitting a smooth monotone curve depicting test performance as a function of the sample size. Learn2Evaluate has several advantages compared to commonly applied performance estimation methodologies. Firstly, a learning curve offers a graphical overview of a learner. This overview assists in assessing the potential benefit of adding training samples and it provides a more complete comparison between learners than performance estimates at a fixed subsample size. Secondly, a learning curve facilitates in estimating the performance at the total sample size rather than a subsample size. Thirdly, Learn2Evaluate allows the computation of a theoretically justified and useful lower confidence bound. Furthermore, this bound may be tightened by performing a bias correction. The benefits of Learn2Evaluate are illustrated by a simulation study and applications to omics data.
ecpc: An R-package for generic co-data models for high-dimensional prediction
May 16, 2022



Abstract:High-dimensional prediction considers data with more variables than samples. Generic research goals are to find the best predictor or to select variables. Results may be improved by exploiting prior information in the form of co-data, providing complementary data not on the samples, but on the variables. We consider adaptive ridge penalised generalised linear and Cox models, in which the variable specific ridge penalties are adapted to the co-data to give a priori more weight to more important variables. The R-package ecpc originally accommodated various and possibly multiple co-data sources, including categorical co-data, i.e. groups of variables, and continuous co-data. Continuous co-data, however, was handled by adaptive discretisation, potentially inefficiently modelling and losing information. Here, we present an extension to the method and software for generic co-data models, particularly for continuous co-data. At the basis lies a classical linear regression model, regressing prior variance weights on the co-data. Co-data variables are then estimated with empirical Bayes moment estimation. After placing the estimation procedure in the classical regression framework, extension to generalised additive and shape constrained co-data models is straightforward. Besides, we show how ridge penalties may be transformed to elastic net penalties with the R-package squeezy. In simulation studies we first compare various co-data models for continuous co-data from the extension to the original method. Secondly, we compare variable selection performance to other variable selection methods. Moreover, we demonstrate use of the package in several examples throughout the paper.
Fast marginal likelihood estimation of penalties for group-adaptive elastic net
Jan 11, 2021



Abstract:Nowadays, clinical research routinely uses omics data, such as gene expression, for predicting clinical outcomes or selecting markers. Additionally, so-called co-data are often available, providing complementary information on the covariates, like p-values from previously published studies or groups of genes corresponding to pathways. Elastic net penalisation is widely used for prediction and covariate selection. Group-adaptive elastic net penalisation learns from co-data to improve the prediction and covariate selection, by penalising important groups of covariates less than other groups. Existing methods are, however, computationally expensive. Here we present a fast method for marginal likelihood estimation of group-adaptive elastic net penalties for generalised linear models. We first derive a low-dimensional representation of the Taylor approximation of the marginal likelihood and its first derivative for group-adaptive ridge penalties, to efficiently estimate these penalties. Then we show by using asymptotic normality of the linear predictors that the marginal likelihood for elastic net models may be approximated well by the marginal likelihood for ridge models. The ridge group penalties are then transformed to elastic net group penalties by using the variance function. The method allows for overlapping groups and unpenalised variables. We demonstrate the method in a model-based simulation study and an application to cancer genomics. The method substantially decreases computation time and outperforms or matches other methods by learning from co-data.
Flexible co-data learning for high-dimensional prediction
May 08, 2020



Abstract:Clinical research often focuses on complex traits in which many variables play a role in mechanisms driving, or curing, diseases. Clinical prediction is hard when data is high-dimensional, but additional information, like domain knowledge and previously published studies, may be helpful to improve predictions. Such complementary data, or co-data, provide information on the covariates, such as genomic location or p-values from external studies. Our method enables exploiting multiple and various co-data sources to improve predictions. We use discrete or continuous co-data to define possibly overlapping or hierarchically structured groups of covariates. These are then used to estimate adaptive multi-group ridge penalties for generalised linear and Cox models. We combine empirical Bayes estimation of group penalty hyperparameters with an extra level of shrinkage. This renders a uniquely flexible framework as any type of shrinkage can be used on the group level. The hyperparameter shrinkage learns how relevant a specific co-data source is, counters overfitting of hyperparameters for many groups, and accounts for structured co-data. We describe various types of co-data and propose suitable forms of hypershrinkage. The method is very versatile, as it allows for integration and weighting of multiple co-data sets, inclusion of unpenalised covariates and posterior variable selection. We demonstrate it on two cancer genomics applications and show that it may improve the performance of other dense and parsimonious prognostic models substantially, and stabilises variable selection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge