Co-data Learning for Bayesian Additive Regression Trees
Paper and Code
Nov 16, 2023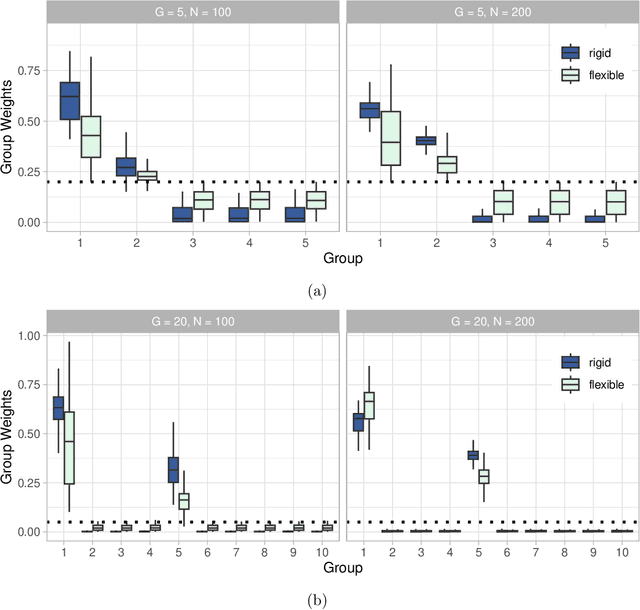
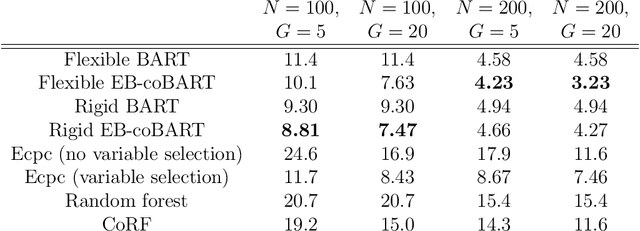
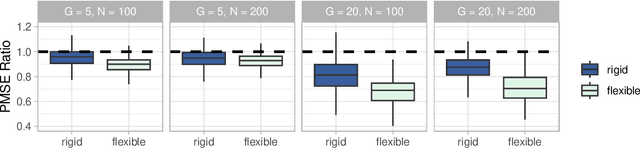
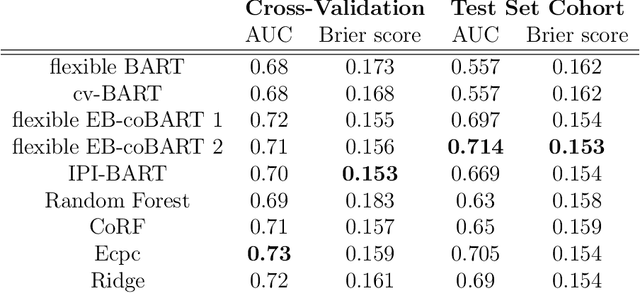
Medical prediction applications often need to deal with small sample sizes compared to the number of covariates. Such data pose problems for prediction and variable selection, especially when the covariate-response relationship is complicated. To address these challenges, we propose to incorporate co-data, i.e. external information on the covariates, into Bayesian additive regression trees (BART), a sum-of-trees prediction model that utilizes priors on the tree parameters to prevent overfitting. To incorporate co-data, an empirical Bayes (EB) framework is developed that estimates, assisted by a co-data model, prior covariate weights in the BART model. The proposed method can handle multiple types of co-data simultaneously. Furthermore, the proposed EB framework enables the estimation of the other hyperparameters of BART as well, rendering an appealing alternative to cross-validation. We show that the method finds relevant covariates and that it improves prediction compared to default BART in simulations. If the covariate-response relationship is nonlinear, the method benefits from the flexibility of BART to outperform regression-based co-data learners. Finally, the use of co-data enhances prediction in an application to diffuse large B-cell lymphoma prognosis based on clinical covariates, gene mutations, DNA translocations, and DNA copy number data. Keywords: Bayesian additive regression trees; Empirical Bayes; Co-data; High-dimensional data; Omics; Prediction
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge