Jeroen M. Goedhart
Co-data Learning for Bayesian Additive Regression Trees
Nov 16, 2023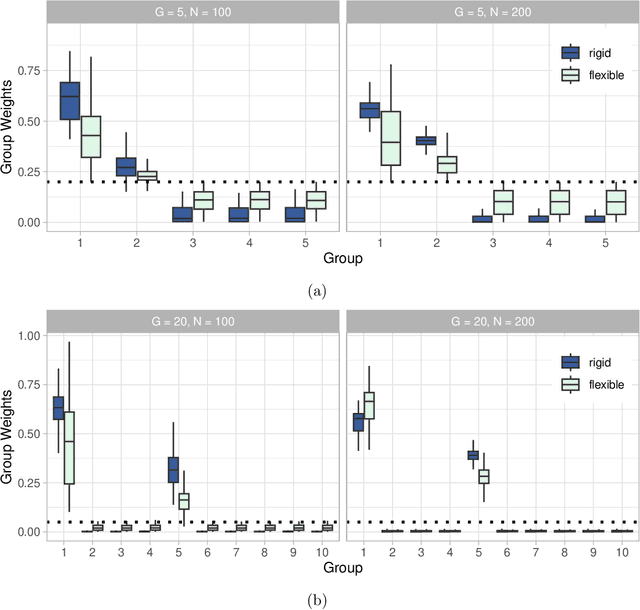
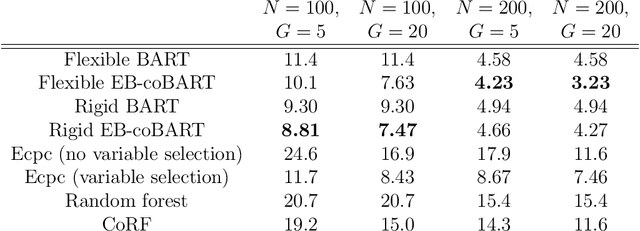
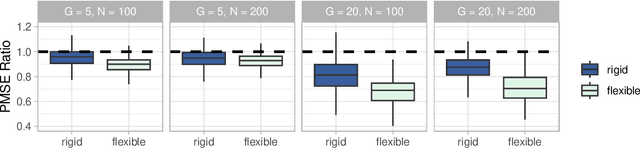
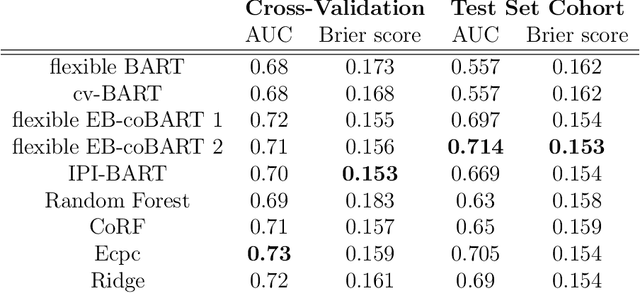
Abstract:Medical prediction applications often need to deal with small sample sizes compared to the number of covariates. Such data pose problems for prediction and variable selection, especially when the covariate-response relationship is complicated. To address these challenges, we propose to incorporate co-data, i.e. external information on the covariates, into Bayesian additive regression trees (BART), a sum-of-trees prediction model that utilizes priors on the tree parameters to prevent overfitting. To incorporate co-data, an empirical Bayes (EB) framework is developed that estimates, assisted by a co-data model, prior covariate weights in the BART model. The proposed method can handle multiple types of co-data simultaneously. Furthermore, the proposed EB framework enables the estimation of the other hyperparameters of BART as well, rendering an appealing alternative to cross-validation. We show that the method finds relevant covariates and that it improves prediction compared to default BART in simulations. If the covariate-response relationship is nonlinear, the method benefits from the flexibility of BART to outperform regression-based co-data learners. Finally, the use of co-data enhances prediction in an application to diffuse large B-cell lymphoma prognosis based on clinical covariates, gene mutations, DNA translocations, and DNA copy number data. Keywords: Bayesian additive regression trees; Empirical Bayes; Co-data; High-dimensional data; Omics; Prediction
Estimation of Predictive Performance in High-Dimensional Data Settings using Learning Curves
Jun 08, 2022



Abstract:In high-dimensional prediction settings, it remains challenging to reliably estimate the test performance. To address this challenge, a novel performance estimation framework is presented. This framework, called Learn2Evaluate, is based on learning curves by fitting a smooth monotone curve depicting test performance as a function of the sample size. Learn2Evaluate has several advantages compared to commonly applied performance estimation methodologies. Firstly, a learning curve offers a graphical overview of a learner. This overview assists in assessing the potential benefit of adding training samples and it provides a more complete comparison between learners than performance estimates at a fixed subsample size. Secondly, a learning curve facilitates in estimating the performance at the total sample size rather than a subsample size. Thirdly, Learn2Evaluate allows the computation of a theoretically justified and useful lower confidence bound. Furthermore, this bound may be tightened by performing a bias correction. The benefits of Learn2Evaluate are illustrated by a simulation study and applications to omics data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge