Mario Wieser
PCIM: Learning Pixel Attributions via Pixel-wise Channel Isolation Mixing in High Content Imaging
Dec 03, 2024Abstract:Deep Neural Networks (DNNs) have shown remarkable success in various computer vision tasks. However, their black-box nature often leads to difficulty in interpreting their decisions, creating an unfilled need for methods to explain the decisions, and ultimately forming a barrier to their wide acceptance especially in biomedical applications. This work introduces a novel method, Pixel-wise Channel Isolation Mixing (PCIM), to calculate pixel attribution maps, highlighting the image parts most crucial for a classification decision but without the need to extract internal network states or gradients. Unlike existing methods, PCIM treats each pixel as a distinct input channel and trains a blending layer to mix these pixels, reflecting specific classifications. This unique approach allows the generation of pixel attribution maps for each image, but agnostic to the choice of the underlying classification network. Benchmark testing on three application relevant, diverse high content Imaging datasets show state-of-the-art performance, particularly for model fidelity and localization ability in both, fluorescence and bright field High Content Imaging. PCIM contributes as a unique and effective method for creating pixel-level attribution maps from arbitrary DNNs, enabling interpretability and trust.
Learning Channel Importance for High Content Imaging with Interpretable Deep Input Channel Mixing
Aug 31, 2023Abstract:Uncovering novel drug candidates for treating complex diseases remain one of the most challenging tasks in early discovery research. To tackle this challenge, biopharma research established a standardized high content imaging protocol that tags different cellular compartments per image channel. In order to judge the experimental outcome, the scientist requires knowledge about the channel importance with respect to a certain phenotype for decoding the underlying biology. In contrast to traditional image analysis approaches, such experiments are nowadays preferably analyzed by deep learning based approaches which, however, lack crucial information about the channel importance. To overcome this limitation, we present a novel approach which utilizes multi-spectral information of high content images to interpret a certain aspect of cellular biology. To this end, we base our method on image blending concepts with alpha compositing for an arbitrary number of channels. More specifically, we introduce DCMIX, a lightweight, scaleable and end-to-end trainable mixing layer which enables interpretable predictions in high content imaging while retaining the benefits of deep learning based methods. We employ an extensive set of experiments on both MNIST and RXRX1 datasets, demonstrating that DCMIX learns the biologically relevant channel importance without scarifying prediction performance.
Learning Conditional Invariance through Cycle Consistency
Nov 25, 2021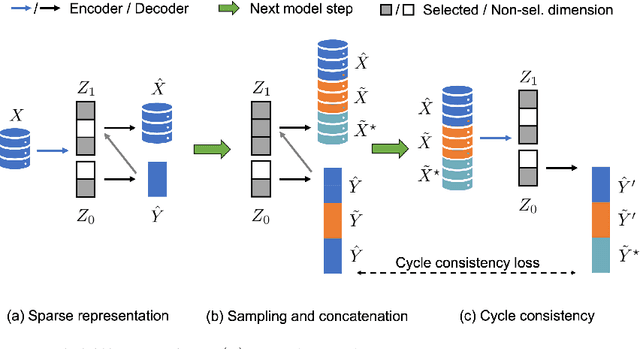
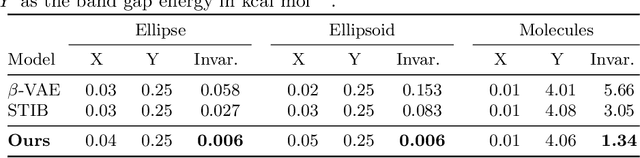
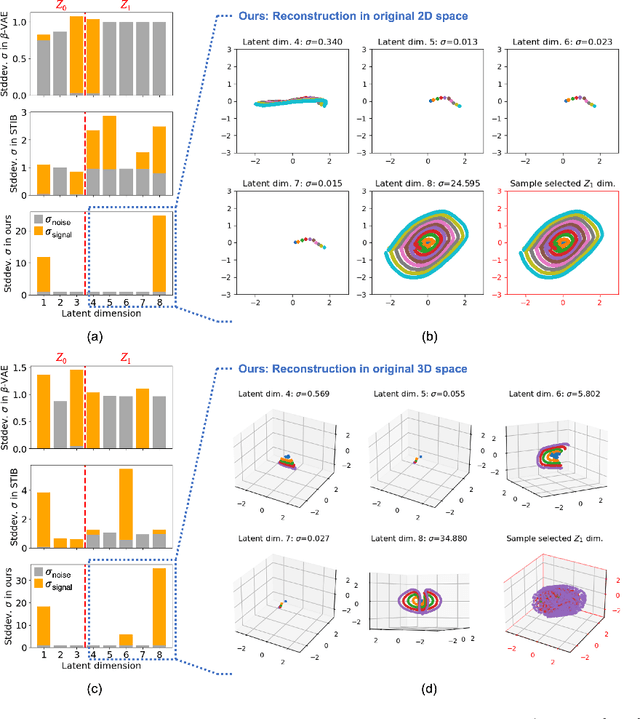
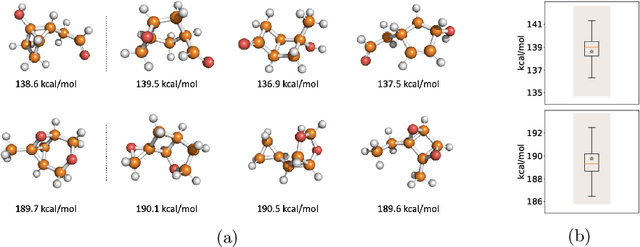
Abstract:Identifying meaningful and independent factors of variation in a dataset is a challenging learning task frequently addressed by means of deep latent variable models. This task can be viewed as learning symmetry transformations preserving the value of a chosen property along latent dimensions. However, existing approaches exhibit severe drawbacks in enforcing the invariance property in the latent space. We address these shortcomings with a novel approach to cycle consistency. Our method involves two separate latent subspaces for the target property and the remaining input information, respectively. In order to enforce invariance as well as sparsity in the latent space, we incorporate semantic knowledge by using cycle consistency constraints relying on property side information. The proposed method is based on the deep information bottleneck and, in contrast to other approaches, allows using continuous target properties and provides inherent model selection capabilities. We demonstrate on synthetic and molecular data that our approach identifies more meaningful factors which lead to sparser and more interpretable models with improved invariance properties.
3DMolNet: A Generative Network for Molecular Structures
Oct 08, 2020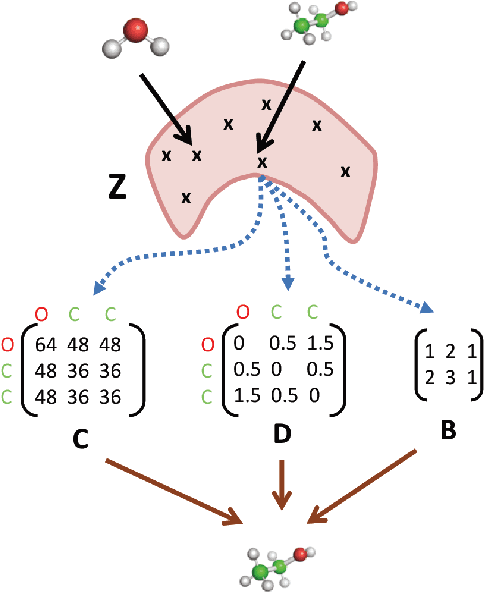
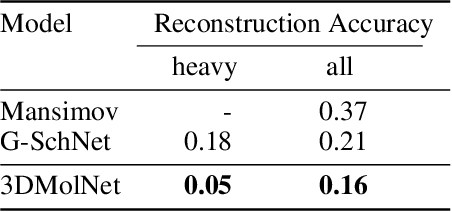
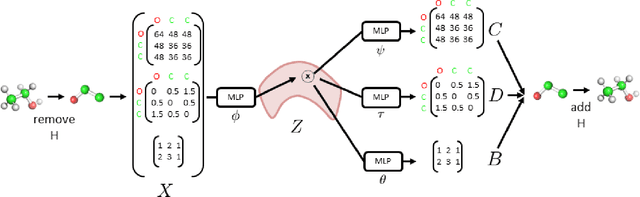
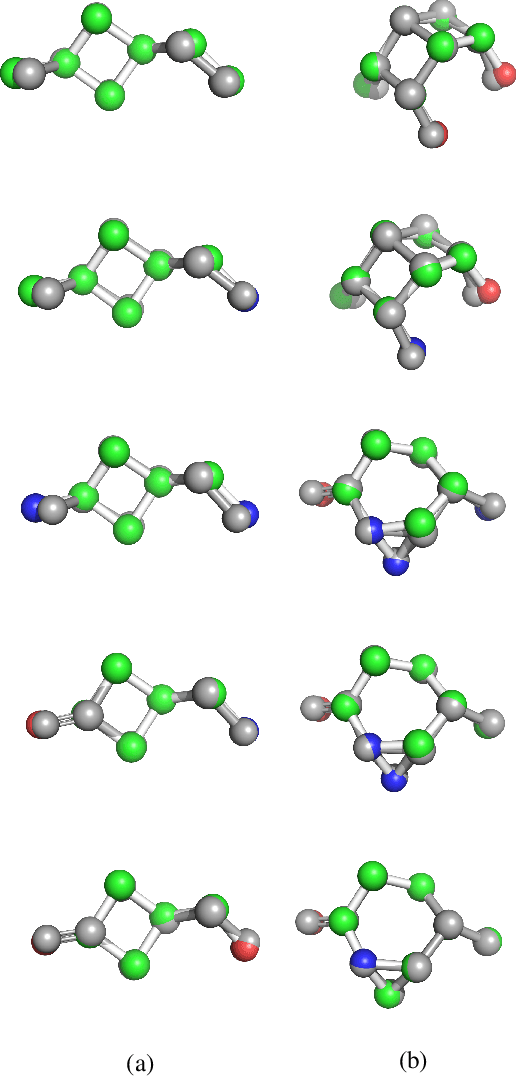
Abstract:With the recent advances in machine learning for quantum chemistry, it is now possible to predict the chemical properties of compounds and to generate novel molecules. Existing generative models mostly use a string- or graph-based representation, but the precise three-dimensional coordinates of the atoms are usually not encoded. First attempts in this direction have been proposed, where autoregressive or GAN-based models generate atom coordinates. Those either lack a latent space in the autoregressive setting, such that a smooth exploration of the compound space is not possible, or cannot generalize to varying chemical compositions. We propose a new approach to efficiently generate molecular structures that are not restricted to a fixed size or composition. Our model is based on the variational autoencoder which learns a translation-, rotation-, and permutation-invariant low-dimensional representation of molecules. Our experiments yield a mean reconstruction error below 0.05 Angstrom, outperforming the current state-of-the-art methods by a factor of four, and which is even lower than the spatial quantization error of most chemical descriptors. The compositional and structural validity of newly generated molecules has been confirmed by quantum chemical methods in a set of experiments.
Inverse Learning of Symmetry Transformations
Feb 07, 2020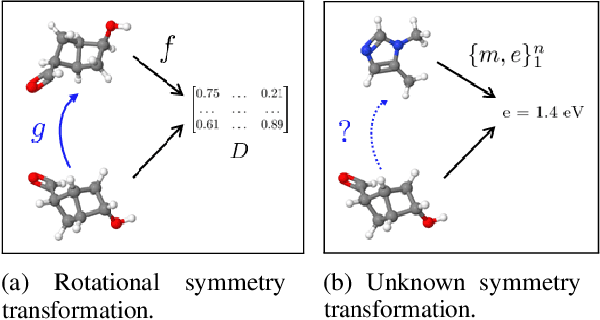
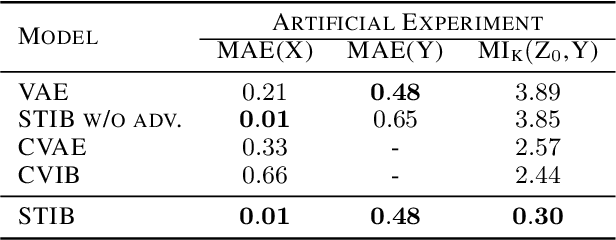
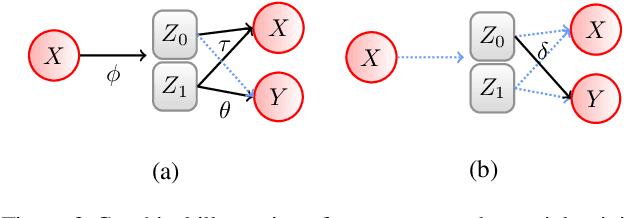
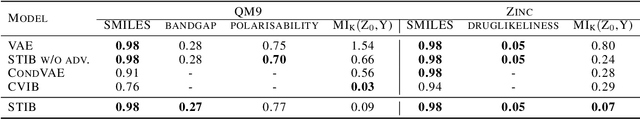
Abstract:Symmetry transformations induce invariances and are a crucial building block of modern machine learning algorithms. Some transformations can be described analytically, e.g. geometric invariances. However, in many complex domains, such as the chemical space, invariances can be observed yet the corresponding symmetry transformation cannot be formulated analytically. Thus, the goal of our work is to learn the symmetry transformation that induced this invariance. To address this task, we propose learning two latent subspaces, where the first subspace captures the property and the second subspace the remaining invariant information. Our approach is based on the deep information bottleneck principle in combination with a mutual information regulariser. Unlike previous methods however, we focus on estimating mutual information in continuous rather than binary settings. This poses many challenges as mutual information cannot be meaningfully minimised in continuous domains. Therefore, we base the calculation of mutual information on correlation matrices in combination with a bijective variable transformation. Extensive experiments demonstrate that our model outperforms state-of-the-art methods on artificial and molecular datasets.
Learning Extremal Representations with Deep Archetypal Analysis
Feb 03, 2020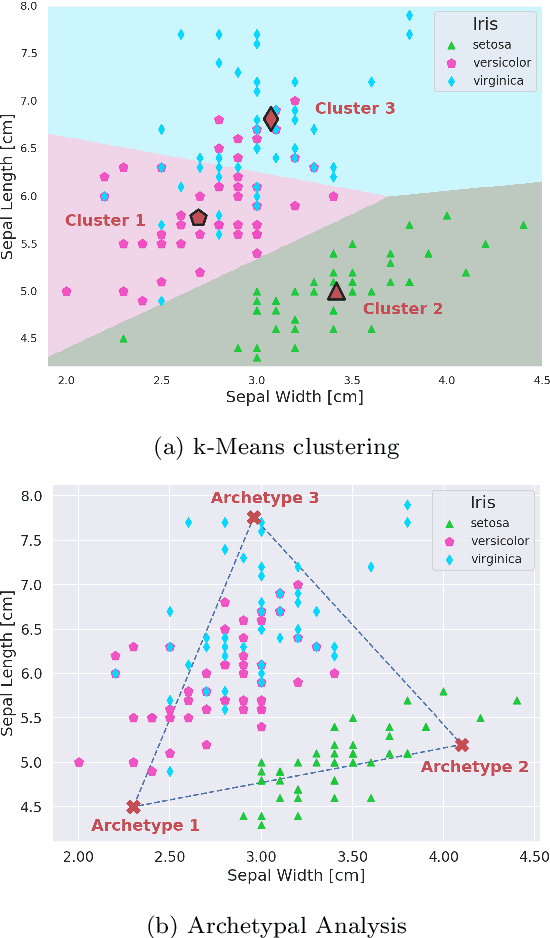
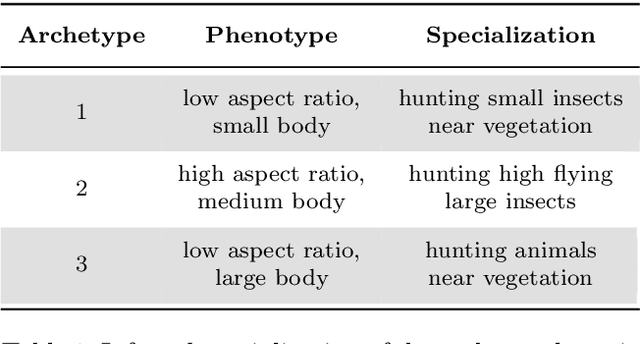
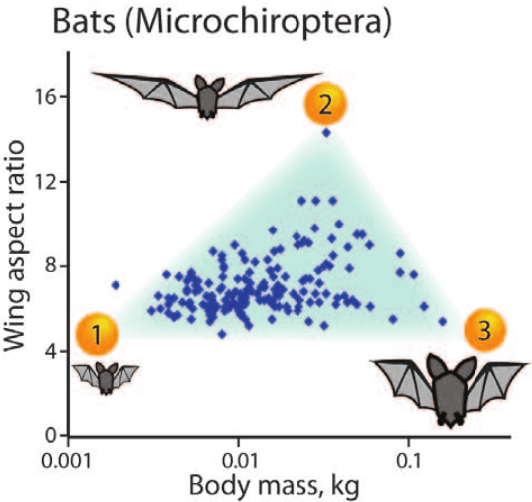
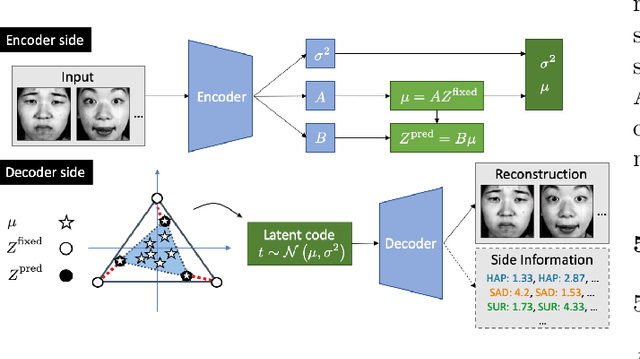
Abstract:Archetypes are typical population representatives in an extremal sense, where typicality is understood as the most extreme manifestation of a trait or feature. In linear feature space, archetypes approximate the data convex hull allowing all data points to be expressed as convex mixtures of archetypes. However, it might not always be possible to identify meaningful archetypes in a given feature space. Learning an appropriate feature space and identifying suitable archetypes simultaneously addresses this problem. This paper introduces a generative formulation of the linear archetype model, parameterized by neural networks. By introducing the distance-dependent archetype loss, the linear archetype model can be integrated into the latent space of a variational autoencoder, and an optimal representation with respect to the unknown archetypes can be learned end-to-end. The reformulation of linear Archetypal Analysis as deep variational information bottleneck, allows the incorporation of arbitrarily complex side information during training. Furthermore, an alternative prior, based on a modified Dirichlet distribution, is proposed. The real-world applicability of the proposed method is demonstrated by exploring archetypes of female facial expressions while using multi-rater based emotion scores of these expressions as side information. A second application illustrates the exploration of the chemical space of small organic molecules. In this experiment, it is demonstrated that exchanging the side information but keeping the same set of molecules, e. g. using as side information the heat capacity of each molecule instead of the band gap energy, will result in the identification of different archetypes. As an application, these learned representations of chemical space might reveal distinct starting points for de novo molecular design.
Deep Archetypal Analysis
Jan 30, 2019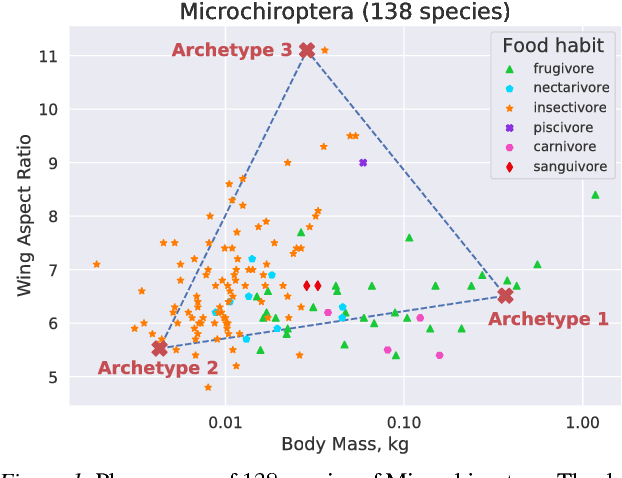
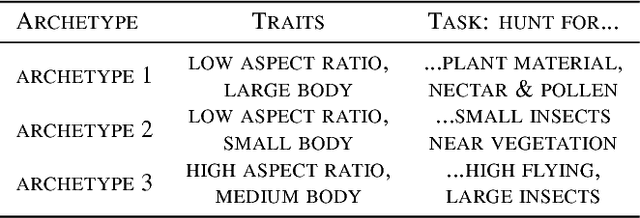

Abstract:"Deep Archetypal Analysis" generates latent representations of high-dimensional datasets in terms of fractions of intuitively understandable basic entities called archetypes. The proposed method is an extension of linear "Archetypal Analysis" (AA), an unsupervised method to represent multivariate data points as sparse convex combinations of extremal elements of the dataset. Unlike the original formulation of AA, "Deep AA" can also handle side information and provides the ability for data-driven representation learning which reduces the dependence on expert knowledge. Our method is motivated by studies of evolutionary trade-offs in biology where archetypes are species highly adapted to a single task. Along these lines, we demonstrate that "Deep AA" also lends itself to the supervised exploration of chemical space, marking a distinct starting point for de novo molecular design. In the unsupervised setting we show how "Deep AA" is used on CelebA to identify archetypal faces. These can then be superimposed in order to generate new faces which inherit dominant traits of the archetypes they are based on.
Informed MCMC with Bayesian Neural Networks for Facial Image Analysis
Nov 29, 2018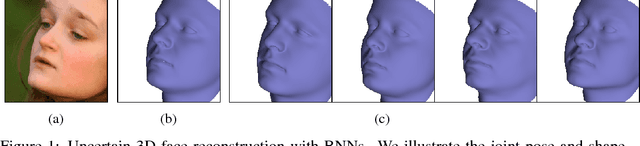
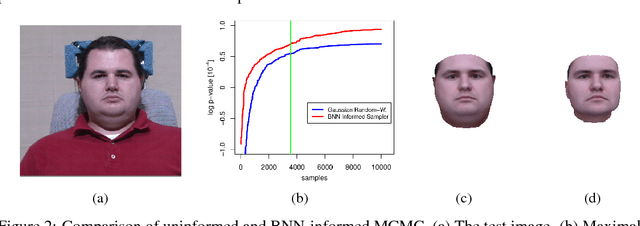
Abstract:Computer vision tasks are difficult because of the large variability in the data that is induced by changes in light, background, partial occlusion as well as the varying pose, texture, and shape of objects. Generative approaches to computer vision allow us to overcome this difficulty by explicitly modeling the physical image formation process. Using generative object models, the analysis of an observed image is performed via Bayesian inference of the posterior distribution. This conceptually simple approach tends to fail in practice because of several difficulties stemming from sampling the posterior distribution: high-dimensionality and multi-modality of the posterior distribution as well as expensive simulation of the rendering process. The main difficulty of sampling approaches in a computer vision context is choosing the proposal distribution accurately so that maxima of the posterior are explored early and the algorithm quickly converges to a valid image interpretation. In this work, we propose to use a Bayesian Neural Network for estimating an image dependent proposal distribution. Compared to a standard Gaussian random walk proposal, this accelerates the sampler in finding regions of the posterior with high value. In this way, we can significantly reduce the number of samples needed to perform facial image analysis.
Estimating Causal Effects With Partial Covariates For Clinical Interpretability
Nov 26, 2018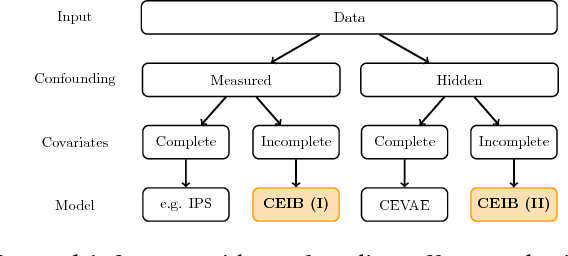
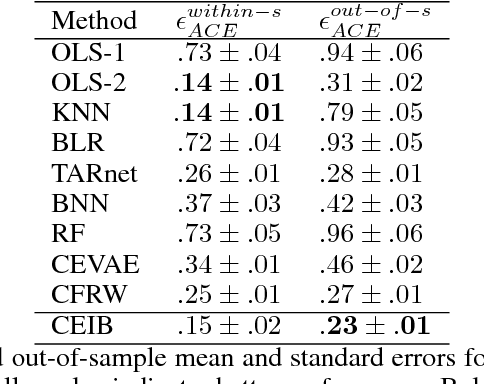


Abstract:Estimating the causal effects of an intervention in the presence of confounding is a frequently occurring problem in applications such as medicine. The task is challenging since there may be multiple confounding factors, some of which may be missing, and inferences must be made from high-dimensional, noisy measurements. In this paper, we propose a decision-theoretic approach to estimate the causal effects of interventions where a subset of the covariates is unavailable for some patients during testing. Our approach uses the information bottleneck principle to perform a discrete, low-dimensional sufficient reduction of the covariate data to estimate a distribution over confounders. In doing so, we can estimate the causal effect of an intervention where only partial covariate information is available. Our results on a causal inference benchmark and a real application for treating sepsis show that our method achieves state-of-the-art performance, without sacrificing interpretability.
Cause-Effect Deep Information Bottleneck For Incomplete Covariates
Oct 08, 2018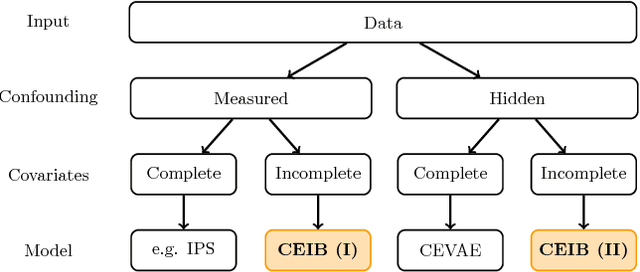


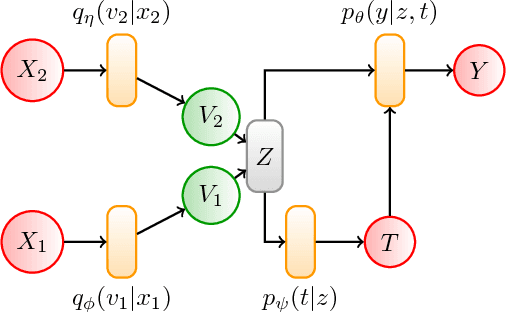
Abstract:Estimating the causal effects of an intervention in the presence of confounding is a frequently occurring problem in applications such as medicine. The task is challenging since there may be multiple confounding factors, some of which may be missing, and inferences must be made from high-dimensional, noisy measurements. In this paper, we propose a decision-theoretic approach to estimate the causal effects of interventions where a subset of the covariates is unavailable for some patients during testing. Our approach uses the information bottleneck principle to perform a discrete, low-dimensional sufficient reduction of the covariate data to estimate a distribution over confounders. In doing so, we can estimate the causal effect of an intervention where only partial covariate information is available. Our results on a causal inference benchmark and a real application for treating sepsis show that our method achieves state-of-the-art performance, without sacrificing interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge