Maria Mahbub
ORCHID: Orchestrated Retrieval-Augmented Classification with Human-in-the-Loop Intelligent Decision-Making for High-Risk Property
Nov 07, 2025


Abstract:High-Risk Property (HRP) classification is critical at U.S. Department of Energy (DOE) sites, where inventories include sensitive and often dual-use equipment. Compliance must track evolving rules designated by various export control policies to make transparent and auditable decisions. Traditional expert-only workflows are time-consuming, backlog-prone, and struggle to keep pace with shifting regulatory boundaries. We demo ORCHID, a modular agentic system for HRP classification that pairs retrieval-augmented generation (RAG) with human oversight to produce policy-based outputs that can be audited. Small cooperating agents, retrieval, description refiner, classifier, validator, and feedback logger, coordinate via agent-to-agent messaging and invoke tools through the Model Context Protocol (MCP) for model-agnostic on-premise operation. The interface follows an Item to Evidence to Decision loop with step-by-step reasoning, on-policy citations, and append-only audit bundles (run-cards, prompts, evidence). In preliminary tests on real HRP cases, ORCHID improves accuracy and traceability over a non-agentic baseline while deferring uncertain items to Subject Matter Experts (SMEs). The demonstration shows single item submission, grounded citations, SME feedback capture, and exportable audit artifacts, illustrating a practical path to trustworthy LLM assistance in sensitive DOE compliance workflows.
Advancing NLP Security by Leveraging LLMs as Adversarial Engines
Oct 23, 2024Abstract:This position paper proposes a novel approach to advancing NLP security by leveraging Large Language Models (LLMs) as engines for generating diverse adversarial attacks. Building upon recent work demonstrating LLMs' effectiveness in creating word-level adversarial examples, we argue for expanding this concept to encompass a broader range of attack types, including adversarial patches, universal perturbations, and targeted attacks. We posit that LLMs' sophisticated language understanding and generation capabilities can produce more effective, semantically coherent, and human-like adversarial examples across various domains and classifier architectures. This paradigm shift in adversarial NLP has far-reaching implications, potentially enhancing model robustness, uncovering new vulnerabilities, and driving innovation in defense mechanisms. By exploring this new frontier, we aim to contribute to the development of more secure, reliable, and trustworthy NLP systems for critical applications.
Hiding-in-Plain-Sight (HiPS) Attack on CLIP for Targetted Object Removal from Images
Oct 16, 2024
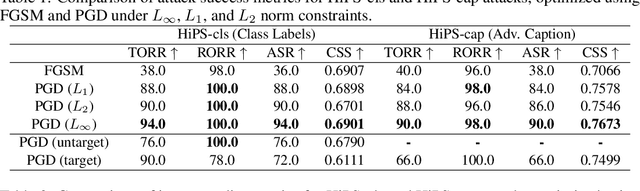


Abstract:Machine learning models are known to be vulnerable to adversarial attacks, but traditional attacks have mostly focused on single-modalities. With the rise of large multi-modal models (LMMs) like CLIP, which combine vision and language capabilities, new vulnerabilities have emerged. However, prior work in multimodal targeted attacks aim to completely change the model's output to what the adversary wants. In many realistic scenarios, an adversary might seek to make only subtle modifications to the output, so that the changes go unnoticed by downstream models or even by humans. We introduce Hiding-in-Plain-Sight (HiPS) attacks, a novel class of adversarial attacks that subtly modifies model predictions by selectively concealing target object(s), as if the target object was absent from the scene. We propose two HiPS attack variants, HiPS-cls and HiPS-cap, and demonstrate their effectiveness in transferring to downstream image captioning models, such as CLIP-Cap, for targeted object removal from image captions.
Leveraging Large Language Models to Extract Information on Substance Use Disorder Severity from Clinical Notes: A Zero-shot Learning Approach
Mar 18, 2024



Abstract:Substance use disorder (SUD) poses a major concern due to its detrimental effects on health and society. SUD identification and treatment depend on a variety of factors such as severity, co-determinants (e.g., withdrawal symptoms), and social determinants of health. Existing diagnostic coding systems used by American insurance providers, like the International Classification of Diseases (ICD-10), lack granularity for certain diagnoses, but clinicians will add this granularity (as that found within the Diagnostic and Statistical Manual of Mental Disorders classification or DSM-5) as supplemental unstructured text in clinical notes. Traditional natural language processing (NLP) methods face limitations in accurately parsing such diverse clinical language. Large Language Models (LLMs) offer promise in overcoming these challenges by adapting to diverse language patterns. This study investigates the application of LLMs for extracting severity-related information for various SUD diagnoses from clinical notes. We propose a workflow employing zero-shot learning of LLMs with carefully crafted prompts and post-processing techniques. Through experimentation with Flan-T5, an open-source LLM, we demonstrate its superior recall compared to the rule-based approach. Focusing on 11 categories of SUD diagnoses, we show the effectiveness of LLMs in extracting severity information, contributing to improved risk assessment and treatment planning for SUD patients.
Question-Answering System Extracts Information on Injection Drug Use from Clinical Progress Notes
May 15, 2023Abstract:Injection drug use (IDU) is a dangerous health behavior that increases mortality and morbidity. Identifying IDU early and initiating harm reduction interventions can benefit individuals at risk. However, extracting IDU behaviors from patients' electronic health records (EHR) is difficult because there is no International Classification of Disease (ICD) code and the only place IDU information can be indicated are unstructured free-text clinical progress notes. Although natural language processing (NLP) can efficiently extract this information from unstructured data, there are no validated tools. To address this gap in clinical information, we design and demonstrate a question-answering (QA) framework to extract information on IDU from clinical progress notes. Unlike other methods discussed in the literature, the QA model is able to extract various types of information without being constrained by predefined entities, relations, or concepts. Our framework involves two main steps: (1) generating a gold-standard QA dataset and (2) developing and testing the QA model. This paper also demonstrates the QA model's ability to extract IDU-related information on temporally out-of-distribution data. The results indicate that the majority (51%) of the extracted information by the QA model exactly matches the gold-standard answer and 73% of them contain the gold-standard answer with some additional surrounding words.
BioADAPT-MRC: Adversarial Learning-based Domain Adaptation Improves Biomedical Machine Reading Comprehension Task
Feb 26, 2022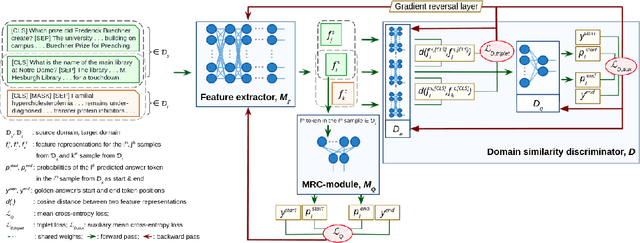

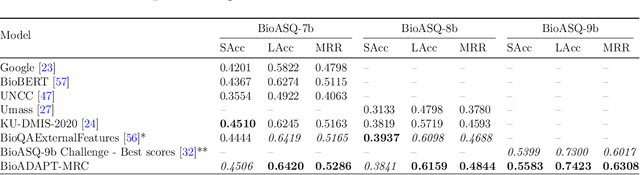
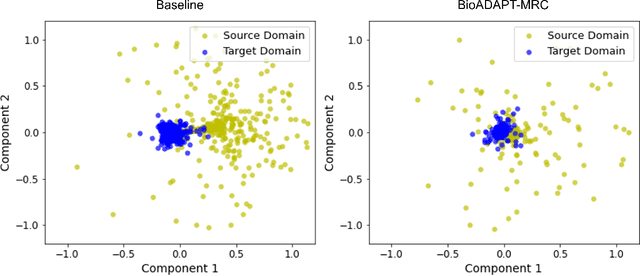
Abstract:Motivation: Biomedical machine reading comprehension (biomedical-MRC) aims to comprehend complex biomedical narratives and assist healthcare professionals in retrieving information from them. The high performance of modern neural network-based MRC systems depends on high-quality, large-scale, human-annotated training datasets. In the biomedical domain, a crucial challenge in creating such datasets is the requirement for domain knowledge, inducing the scarcity of labeled data and the need for transfer learning from the labeled general-purpose (source) domain to the biomedical (target) domain. However, there is a discrepancy in marginal distributions between the general-purpose and biomedical domains due to the variances in topics. Therefore, direct-transferring of learned representations from a model trained on a general-purpose domain to the biomedical domain can hurt the model's performance. Results: We present an adversarial learning-based domain adaptation framework for the biomedical machine reading comprehension task (BioADAPT-MRC), a neural network-based method to address the discrepancies in the marginal distributions between the general and biomedical domain datasets. BioADAPT-MRC relaxes the need for generating pseudo labels for training a well-performing biomedical-MRC model. We extensively evaluate the performance of BioADAPT-MRC by comparing it with the best existing methods on three widely used benchmark biomedical-MRC datasets -- BioASQ-7b, BioASQ-8b, and BioASQ-9b. Our results suggest that without using any synthetic or human-annotated data from the biomedical domain, BioADAPT-MRC can achieve state-of-the-art performance on these datasets. Availability: BioADAPT-MRC is freely available as an open-source project at\\https://github.com/mmahbub/BioADAPT-MRC
The Sensitivity of Word Embeddings-based Author Detection Models to Semantic-preserving Adversarial Perturbations
Feb 23, 2021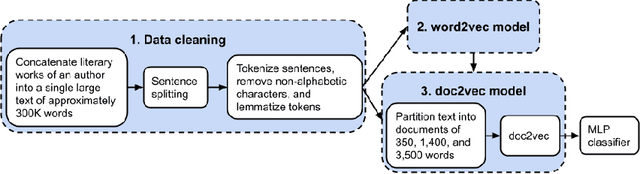

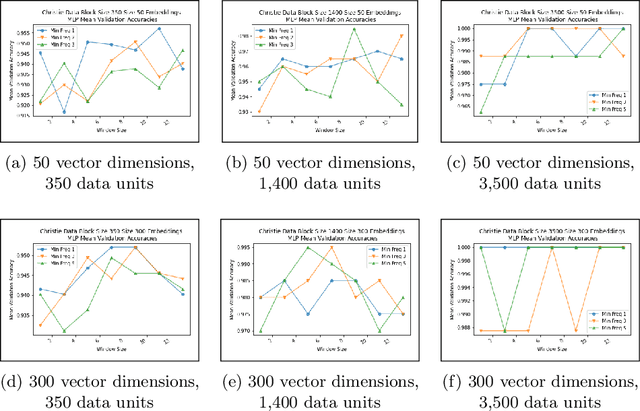

Abstract:Authorship analysis is an important subject in the field of natural language processing. It allows the detection of the most likely writer of articles, news, books, or messages. This technique has multiple uses in tasks related to authorship attribution, detection of plagiarism, style analysis, sources of misinformation, etc. The focus of this paper is to explore the limitations and sensitiveness of established approaches to adversarial manipulations of inputs. To this end, and using those established techniques, we first developed an experimental frame-work for author detection and input perturbations. Next, we experimentally evaluated the performance of the authorship detection model to a collection of semantic-preserving adversarial perturbations of input narratives. Finally, we compare and analyze the effects of different perturbation strategies, input and model configurations, and the effects of these on the author detection model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge