Marc Berndl
It's easy to fool yourself: Case studies on identifying bias and confounding in bio-medical datasets
Dec 12, 2019
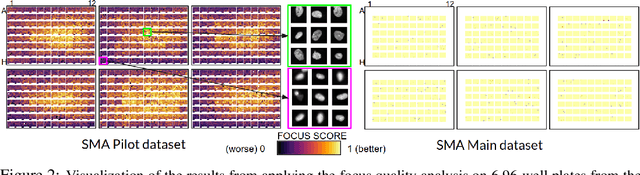
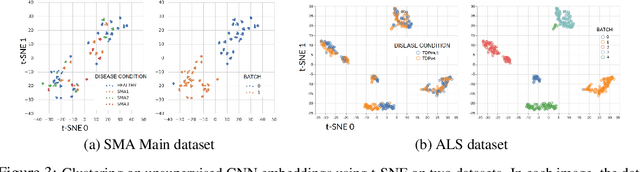
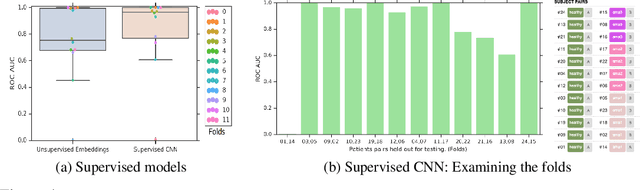
Abstract:Confounding variables are a well known source of nuisance in biomedical studies. They present an even greater challenge when we combine them with black-box machine learning techniques that operate on raw data. This work presents two case studies. In one, we discovered biases arising from systematic errors in the data generation process. In the other, we found a spurious source of signal unrelated to the prediction task at hand. In both cases, our prediction models performed well but under careful examination hidden confounders and biases were revealed. These are cautionary tales on the limits of using machine learning techniques on raw data from scientific experiments.
Molecular Graph Convolutions: Moving Beyond Fingerprints
Aug 18, 2016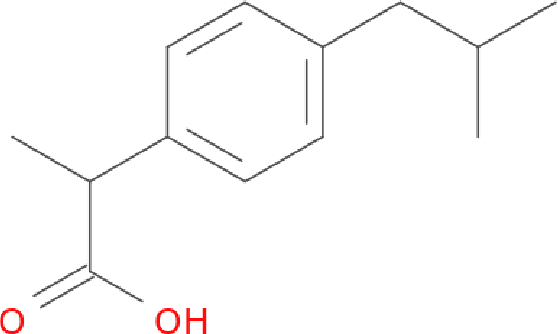
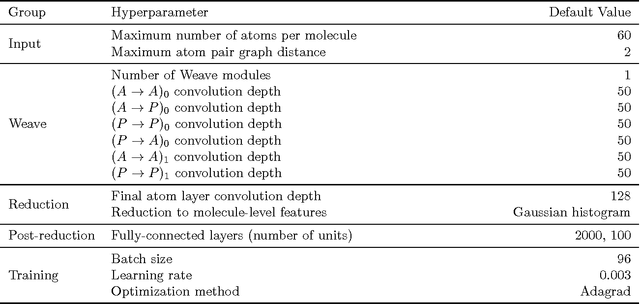
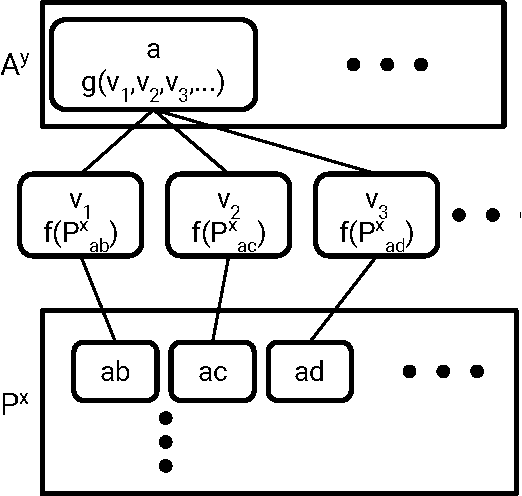
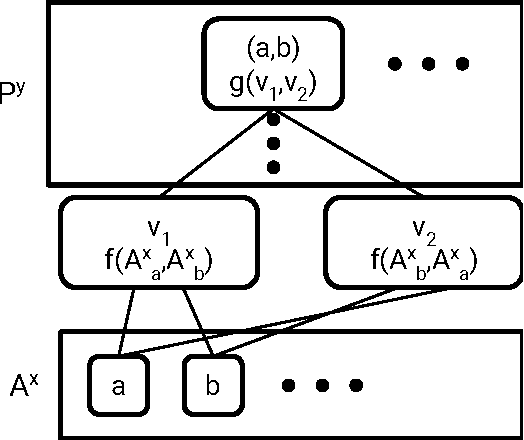
Abstract:Molecular "fingerprints" encoding structural information are the workhorse of cheminformatics and machine learning in drug discovery applications. However, fingerprint representations necessarily emphasize particular aspects of the molecular structure while ignoring others, rather than allowing the model to make data-driven decisions. We describe molecular "graph convolutions", a machine learning architecture for learning from undirected graphs, specifically small molecules. Graph convolutions use a simple encoding of the molecular graph---atoms, bonds, distances, etc.---which allows the model to take greater advantage of information in the graph structure. Although graph convolutions do not outperform all fingerprint-based methods, they (along with other graph-based methods) represent a new paradigm in ligand-based virtual screening with exciting opportunities for future improvement.
* See "Version information" section
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge