Longtian Bao
Single-Pass Document Scanning for Question Answering
Apr 04, 2025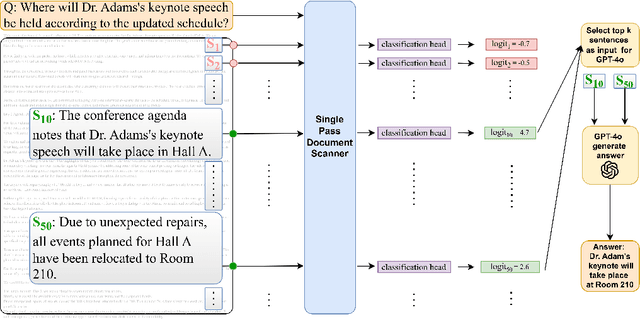
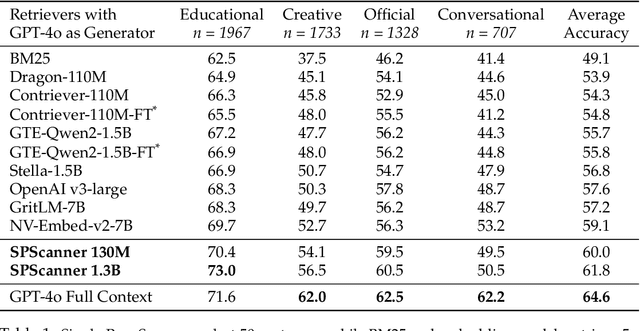
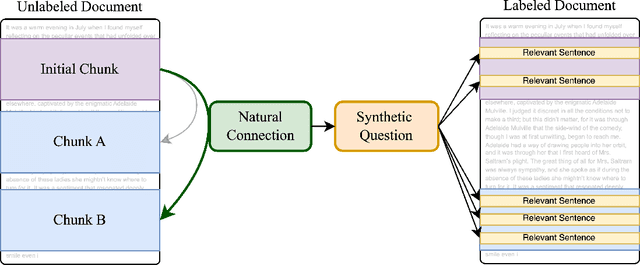
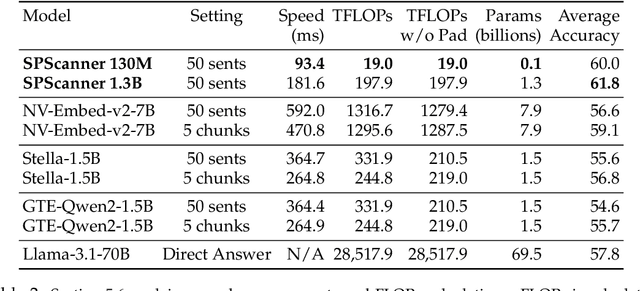
Abstract:Handling extremely large documents for question answering is challenging: chunk-based embedding methods often lose track of important global context, while full-context transformers can be prohibitively expensive for hundreds of thousands of tokens. We propose a single-pass document scanning approach that processes the entire text in linear time, preserving global coherence while deciding which sentences are most relevant to the query. On 41 QA benchmarks, our single-pass scanner consistently outperforms chunk-based embedding methods and competes with large language models at a fraction of the computational cost. By conditioning on the entire preceding context without chunk breaks, the method preserves global coherence, which is especially important for long documents. Overall, single-pass document scanning offers a simple solution for question answering over massive text. All code, datasets, and model checkpoints are available at https://github.com/MambaRetriever/MambaRetriever
Measuring Risk of Bias in Biomedical Reports: The RoBBR Benchmark
Nov 28, 2024



Abstract:Systems that answer questions by reviewing the scientific literature are becoming increasingly feasible. To draw reliable conclusions, these systems should take into account the quality of available evidence, placing more weight on studies that use a valid methodology. We present a benchmark for measuring the methodological strength of biomedical papers, drawing on the risk-of-bias framework used for systematic reviews. The four benchmark tasks, drawn from more than 500 papers, cover the analysis of research study methodology, followed by evaluation of risk of bias in these studies. The benchmark contains 2000 expert-generated bias annotations, and a human-validated pipeline for fine-grained alignment with research paper content. We evaluate a range of large language models on the benchmark, and find that these models fall significantly short of expert-level performance. By providing a standardized tool for measuring judgments of study quality, the benchmark can help to guide systems that perform large-scale aggregation of scientific data. The dataset is available at https://github.com/RoBBR-Benchmark/RoBBR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge