Lizhi He
HeteRAG: A Heterogeneous Retrieval-augmented Generation Framework with Decoupled Knowledge Representations
Apr 12, 2025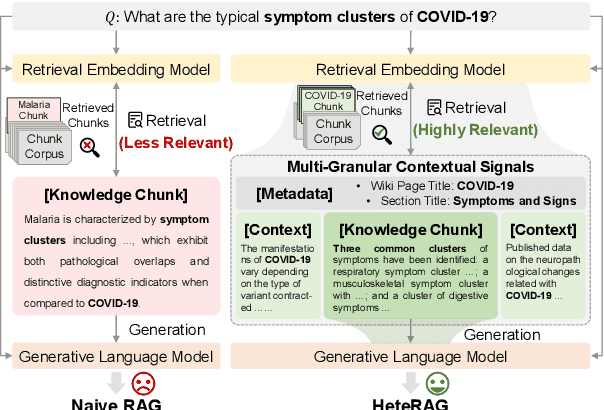

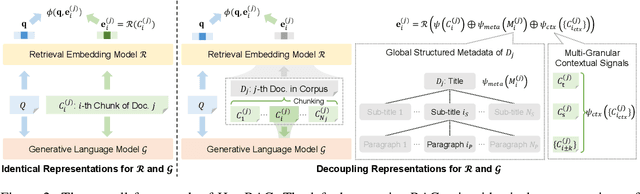

Abstract:Retrieval-augmented generation (RAG) methods can enhance the performance of LLMs by incorporating retrieved knowledge chunks into the generation process. In general, the retrieval and generation steps usually have different requirements for these knowledge chunks. The retrieval step benefits from comprehensive information to improve retrieval accuracy, whereas excessively long chunks may introduce redundant contextual information, thereby diminishing both the effectiveness and efficiency of the generation process. However, existing RAG methods typically employ identical representations of knowledge chunks for both retrieval and generation, resulting in suboptimal performance. In this paper, we propose a heterogeneous RAG framework (\myname) that decouples the representations of knowledge chunks for retrieval and generation, thereby enhancing the LLMs in both effectiveness and efficiency. Specifically, we utilize short chunks to represent knowledge to adapt the generation step and utilize the corresponding chunk with its contextual information from multi-granular views to enhance retrieval accuracy. We further introduce an adaptive prompt tuning method for the retrieval model to adapt the heterogeneous retrieval augmented generation process. Extensive experiments demonstrate that \myname achieves significant improvements compared to baselines.
Citrus: Leveraging Expert Cognitive Pathways in a Medical Language Model for Advanced Medical Decision Support
Feb 26, 2025Abstract:Large language models (LLMs), particularly those with reasoning capabilities, have rapidly advanced in recent years, demonstrating significant potential across a wide range of applications. However, their deployment in healthcare, especially in disease reasoning tasks, is hindered by the challenge of acquiring expert-level cognitive data. In this paper, we introduce Citrus, a medical language model that bridges the gap between clinical expertise and AI reasoning by emulating the cognitive processes of medical experts. The model is trained on a large corpus of simulated expert disease reasoning data, synthesized using a novel approach that accurately captures the decision-making pathways of clinicians. This approach enables Citrus to better simulate the complex reasoning processes involved in diagnosing and treating medical conditions. To further address the lack of publicly available datasets for medical reasoning tasks, we release the last-stage training data, including a custom-built medical diagnostic dialogue dataset. This open-source contribution aims to support further research and development in the field. Evaluations using authoritative benchmarks such as MedQA, covering tasks in medical reasoning and language understanding, show that Citrus achieves superior performance compared to other models of similar size. These results highlight Citrus potential to significantly enhance medical decision support systems, providing a more accurate and efficient tool for clinical decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge