Linghao Feng
PandaGuard: Systematic Evaluation of LLM Safety against Jailbreaking Attacks
May 22, 2025Abstract:Large language models (LLMs) have achieved remarkable capabilities but remain vulnerable to adversarial prompts known as jailbreaks, which can bypass safety alignment and elicit harmful outputs. Despite growing efforts in LLM safety research, existing evaluations are often fragmented, focused on isolated attack or defense techniques, and lack systematic, reproducible analysis. In this work, we introduce PandaGuard, a unified and modular framework that models LLM jailbreak safety as a multi-agent system comprising attackers, defenders, and judges. Our framework implements 19 attack methods and 12 defense mechanisms, along with multiple judgment strategies, all within a flexible plugin architecture supporting diverse LLM interfaces, multiple interaction modes, and configuration-driven experimentation that enhances reproducibility and practical deployment. Built on this framework, we develop PandaBench, a comprehensive benchmark that evaluates the interactions between these attack/defense methods across 49 LLMs and various judgment approaches, requiring over 3 billion tokens to execute. Our extensive evaluation reveals key insights into model vulnerabilities, defense cost-performance trade-offs, and judge consistency. We find that no single defense is optimal across all dimensions and that judge disagreement introduces nontrivial variance in safety assessments. We release the code, configurations, and evaluation results to support transparent and reproducible research in LLM safety.
STEP: A Unified Spiking Transformer Evaluation Platform for Fair and Reproducible Benchmarking
May 16, 2025Abstract:Spiking Transformers have recently emerged as promising architectures for combining the efficiency of spiking neural networks with the representational power of self-attention. However, the lack of standardized implementations, evaluation pipelines, and consistent design choices has hindered fair comparison and principled analysis. In this paper, we introduce \textbf{STEP}, a unified benchmark framework for Spiking Transformers that supports a wide range of tasks, including classification, segmentation, and detection across static, event-based, and sequential datasets. STEP provides modular support for diverse components such as spiking neurons, input encodings, surrogate gradients, and multiple backends (e.g., SpikingJelly, BrainCog). Using STEP, we reproduce and evaluate several representative models, and conduct systematic ablation studies on attention design, neuron types, encoding schemes, and temporal modeling capabilities. We also propose a unified analytical model for energy estimation, accounting for spike sparsity, bitwidth, and memory access, and show that quantized ANNs may offer comparable or better energy efficiency. Our results suggest that current Spiking Transformers rely heavily on convolutional frontends and lack strong temporal modeling, underscoring the need for spike-native architectural innovations. The full code is available at: https://github.com/Fancyssc/STEP
Biologically Inspired Spiking Diffusion Model with Adaptive Lateral Selection Mechanism
Mar 31, 2025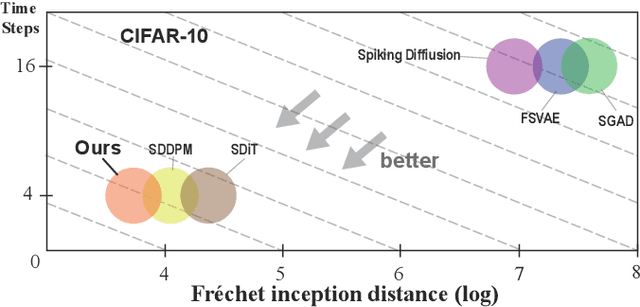

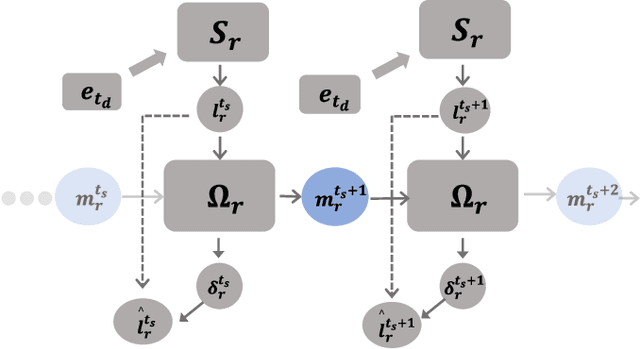

Abstract:Lateral connection is a fundamental feature of biological neural circuits, facilitating local information processing and adaptive learning. In this work, we integrate lateral connections with a substructure selection network to develop a novel diffusion model based on spiking neural networks (SNNs). Unlike conventional artificial neural networks, SNNs employ an intrinsic spiking inner loop to process sequential binary spikes. We leverage this spiking inner loop alongside a lateral connection mechanism to iteratively refine the substructure selection network, enhancing model adaptability and expressivity. Specifically, we design a lateral connection framework comprising a learnable lateral matrix and a lateral mapping function, both implemented using spiking neurons, to dynamically update lateral connections. Through mathematical modeling, we establish that the proposed lateral update mechanism, under a well-defined local objective, aligns with biologically plausible synaptic plasticity principles. Extensive experiments validate the effectiveness of our approach, analyzing the role of substructure selection and lateral connection during training. Furthermore, quantitative comparisons demonstrate that our model consistently surpasses state-of-the-art SNN-based generative models across multiple benchmark datasets.
Time Cell Inspired Temporal Codebook in Spiking Neural Networks for Enhanced Image Generation
May 23, 2024



Abstract:This paper presents a novel approach leveraging Spiking Neural Networks (SNNs) to construct a Variational Quantized Autoencoder (VQ-VAE) with a temporal codebook inspired by hippocampal time cells. This design captures and utilizes temporal dependencies, significantly enhancing the generative capabilities of SNNs. Neuroscientific research has identified hippocampal "time cells" that fire sequentially during temporally structured experiences. Our temporal codebook emulates this behavior by triggering the activation of time cell populations based on similarity measures as input stimuli pass through it. We conducted extensive experiments on standard benchmark datasets, including MNIST, FashionMNIST, CIFAR10, CelebA, and downsampled LSUN Bedroom, to validate our model's performance. Furthermore, we evaluated the effectiveness of the temporal codebook on neuromorphic datasets NMNIST and DVS-CIFAR10, and demonstrated the model's capability with high-resolution datasets such as CelebA-HQ, LSUN Bedroom, and LSUN Church. The experimental results indicate that our method consistently outperforms existing SNN-based generative models across multiple datasets, achieving state-of-the-art performance. Notably, our approach excels in generating high-resolution and temporally consistent data, underscoring the crucial role of temporal information in SNN-based generative modeling.
Neuro-Vision to Language: Image Reconstruction and Language enabled Interaction via Brain Recordings
May 01, 2024



Abstract:Decoding non-invasive brain recordings is crucial for advancing our understanding of human cognition, yet faces challenges from individual differences and complex neural signal representations. Traditional methods require custom models and extensive trials, and lack interpretability in visual reconstruction tasks. Our framework integrating integrates 3D brain structures with visual semantics by Vision Transformer 3D. The unified feature extractor aligns fMRI features with multiple levels of visual embeddings efficiently, removing the need for individual-specific models and allowing extraction from single-trial data. This extractor consolidates multi-level visual features into one network, simplifying integration with Large Language Models (LLMs). Additionally, we have enhanced the fMRI dataset with various fMRI-image related textual data to support multimodal large model development. The integration with LLMs enhances decoding capabilities, enabling tasks like brain captioning, question-answering, detailed descriptions, complex reasoning, and visual reconstruction. Our approach not only shows superior performance across these tasks but also precisely identifies and manipulates language-based concepts within brain signals, enhancing interpretability and providing deeper neural process insights. These advances significantly broaden non-invasive brain decoding applicability in neuroscience and human-computer interaction, setting the stage for advanced brain-computer interfaces and cognitive models.
Spiking Generative Adversarial Network with Attention Scoring Decoding
May 19, 2023



Abstract:Generative models based on neural networks present a substantial challenge within deep learning. As it stands, such models are primarily limited to the domain of artificial neural networks. Spiking neural networks, as the third generation of neural networks, offer a closer approximation to brain-like processing due to their rich spatiotemporal dynamics. However, generative models based on spiking neural networks are not well studied. In this work, we pioneer constructing a spiking generative adversarial network capable of handling complex images. Our first task was to identify the problems of out-of-domain inconsistency and temporal inconsistency inherent in spiking generative adversarial networks. We addressed these issues by incorporating the Earth-Mover distance and an attention-based weighted decoding method, significantly enhancing the performance of our algorithm across several datasets. Experimental results reveal that our approach outperforms existing methods on the MNIST, FashionMNIST, CIFAR10, and CelebA datasets. Moreover, compared with hybrid spiking generative adversarial networks, where the discriminator is an artificial analog neural network, our methodology demonstrates closer alignment with the information processing patterns observed in the mouse.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge