Lina Zhang
Unified Multimodal and Multilingual Retrieval via Multi-Task Learning with NLU Integration
Jan 21, 2026Abstract:Multimodal retrieval systems typically employ Vision Language Models (VLMs) that encode images and text independently into vectors within a shared embedding space. Despite incorporating text encoders, VLMs consistently underperform specialized text models on text-only retrieval tasks. Moreover, introducing additional text encoders increases storage, inference overhead, and exacerbates retrieval inefficiencies, especially in multilingual settings. To address these limitations, we propose a multi-task learning framework that unifies the feature representation across images, long and short texts, and intent-rich queries. To our knowledge, this is the first work to jointly optimize multilingual image retrieval, text retrieval, and natural language understanding (NLU) tasks within a single framework. Our approach integrates image and text retrieval with a shared text encoder that is enhanced by NLU features for intent understanding and retrieval accuracy.
OregairuChar: A Benchmark Dataset for Character Appearance Frequency Analysis in My Teen Romantic Comedy SNAFU
Nov 07, 2025Abstract:The analysis of character appearance frequency is essential for understanding narrative structure, character prominence, and story progression in anime. In this work, we introduce OregairuChar, a benchmark dataset designed for appearance frequency analysis in the anime series My Teen Romantic Comedy SNAFU. The dataset comprises 1600 manually selected frames from the third season, annotated with 2860 bounding boxes across 11 main characters. OregairuChar captures diverse visual challenges, including occlusion, pose variation, and inter-character similarity, providing a realistic basis for appearance-based studies. To enable quantitative research, we benchmark several object detection models on the dataset and leverage their predictions for fine-grained, episode-level analysis of character presence over time. This approach reveals patterns of character prominence and their evolution within the narrative. By emphasizing appearance frequency, OregairuChar serves as a valuable resource for exploring computational narrative dynamics and character-centric storytelling in stylized media.
A Descriptor Is All You Need: Accurate Machine Learning of Nonadiabatic Coupling Vectors
May 29, 2025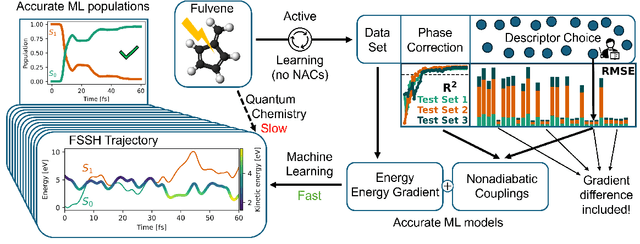
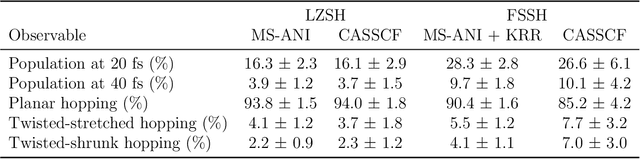
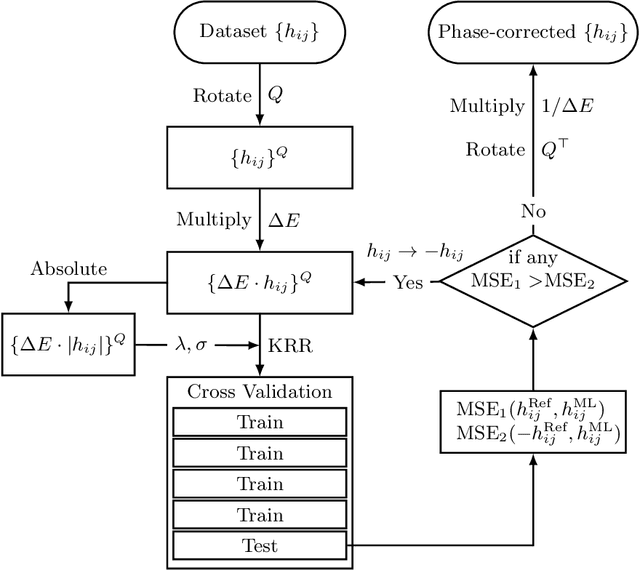
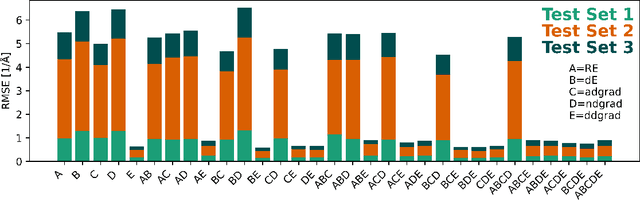
Abstract:Nonadiabatic couplings (NACs) play a crucial role in modeling photochemical and photophysical processes with methods such as the widely used fewest-switches surface hopping (FSSH). There is therefore a strong incentive to machine learn NACs for accelerating simulations. However, this is challenging due to NACs' vectorial, double-valued character and the singularity near a conical intersection seam. For the first time, we design NAC-specific descriptors based on our domain expertise and show that they allow learning NACs with never-before-reported accuracy of $R^2$ exceeding 0.99. The key to success is also our new ML phase-correction procedure. We demonstrate the efficiency and robustness of our approach on a prototypical example of fully ML-driven FSSH simulations of fulvene targeting the SA-2-CASSCF(6,6) electronic structure level. This ML-FSSH dynamics leads to an accurate description of $S_1$ decay while reducing error bars by allowing the execution of a large ensemble of trajectories. Our implementations are available in open-source MLatom.
HiFiSeg: High-Frequency Information Enhanced Polyp Segmentation with Global-Local Vision Transformer
Oct 03, 2024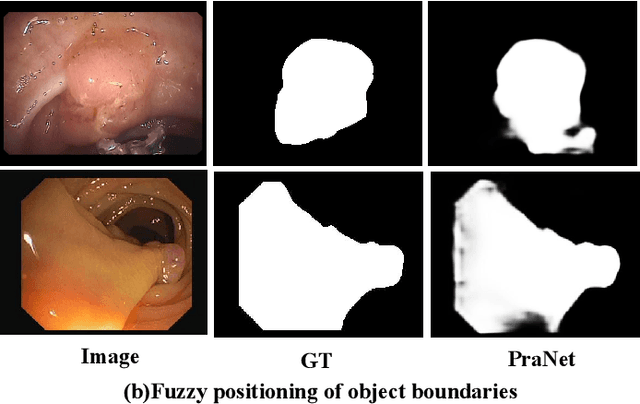
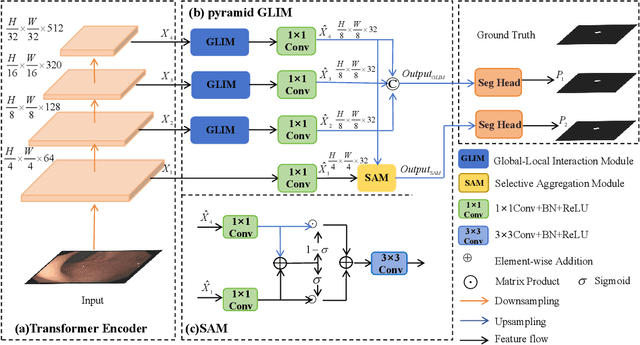
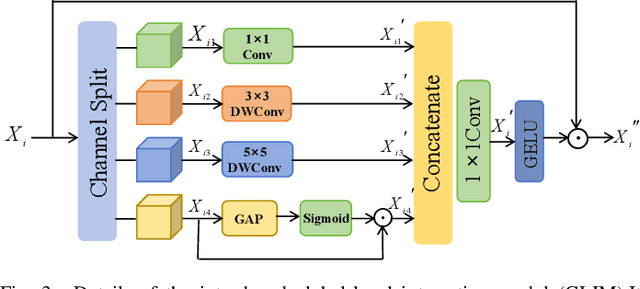
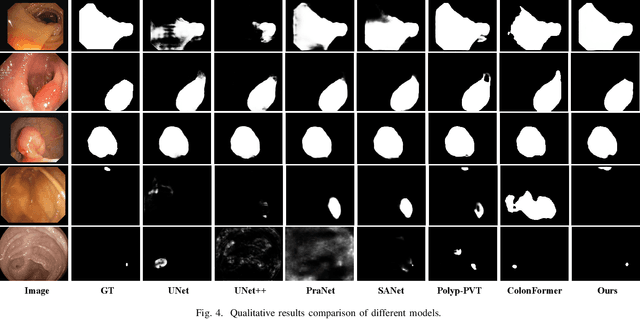
Abstract:Numerous studies have demonstrated the strong performance of Vision Transformer (ViT)-based methods across various computer vision tasks. However, ViT models often struggle to effectively capture high-frequency components in images, which are crucial for detecting small targets and preserving edge details, especially in complex scenarios. This limitation is particularly challenging in colon polyp segmentation, where polyps exhibit significant variability in structure, texture, and shape. High-frequency information, such as boundary details, is essential for achieving precise semantic segmentation in this context. To address these challenges, we propose HiFiSeg, a novel network for colon polyp segmentation that enhances high-frequency information processing through a global-local vision transformer framework. HiFiSeg leverages the pyramid vision transformer (PVT) as its encoder and introduces two key modules: the global-local interaction module (GLIM) and the selective aggregation module (SAM). GLIM employs a parallel structure to fuse global and local information at multiple scales, effectively capturing fine-grained features. SAM selectively integrates boundary details from low-level features with semantic information from high-level features, significantly improving the model's ability to accurately detect and segment polyps. Extensive experiments on five widely recognized benchmark datasets demonstrate the effectiveness of HiFiSeg for polyp segmentation. Notably, the mDice scores on the challenging CVC-ColonDB and ETIS datasets reached 0.826 and 0.822, respectively, underscoring the superior performance of HiFiSeg in handling the specific complexities of this task.
Physics-informed active learning for accelerating quantum chemical simulations
Apr 18, 2024Abstract:Quantum chemical simulations can be greatly accelerated by constructing machine learning potentials, which is often done using active learning (AL). The usefulness of the constructed potentials is often limited by the high effort required and their insufficient robustness in the simulations. Here we introduce the end-to-end AL for constructing robust data-efficient potentials with affordable investment of time and resources and minimum human interference. Our AL protocol is based on the physics-informed sampling of training points, automatic selection of initial data, and uncertainty quantification. The versatility of this protocol is shown in our implementation of quasi-classical molecular dynamics for simulating vibrational spectra, conformer search of a key biochemical molecule, and time-resolved mechanism of the Diels-Alder reaction. These investigations took us days instead of weeks of pure quantum chemical calculations on a high-performance computing cluster.
MLatom 3: Platform for machine learning-enhanced computational chemistry simulations and workflows
Oct 31, 2023
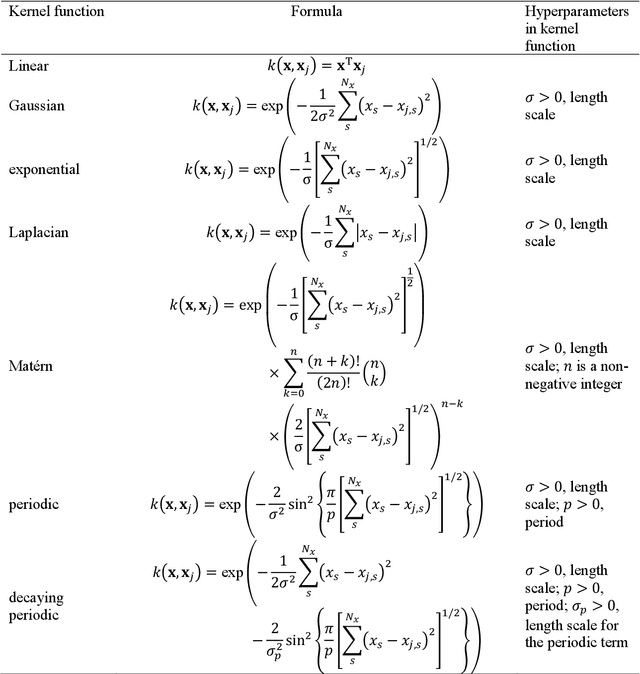
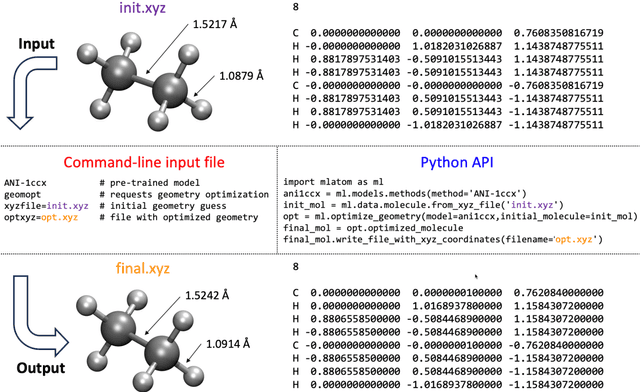
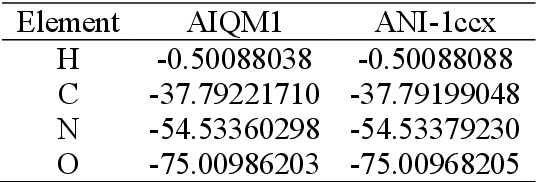
Abstract:Machine learning (ML) is increasingly becoming a common tool in computational chemistry. At the same time, the rapid development of ML methods requires a flexible software framework for designing custom workflows. MLatom 3 is a program package designed to leverage the power of ML to enhance typical computational chemistry simulations and to create complex workflows. This open-source package provides plenty of choice to the users who can run simulations with the command line options, input files, or with scripts using MLatom as a Python package, both on their computers and on the online XACS cloud computing at XACScloud.com. Computational chemists can calculate energies and thermochemical properties, optimize geometries, run molecular and quantum dynamics, and simulate (ro)vibrational, one-photon UV/vis absorption, and two-photon absorption spectra with ML, quantum mechanical, and combined models. The users can choose from an extensive library of methods containing pre-trained ML models and quantum mechanical approximations such as AIQM1 approaching coupled-cluster accuracy. The developers can build their own models using various ML algorithms. The great flexibility of MLatom is largely due to the extensive use of the interfaces to many state-of-the-art software packages and libraries.
Temporal Spatial-Adaptive Interpolation with Deformable Refinement for Electron Microscopic Images
Jan 17, 2021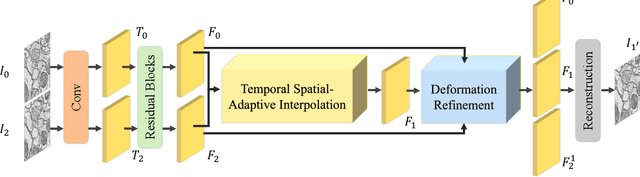
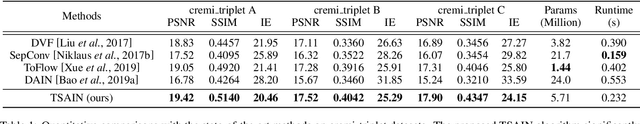
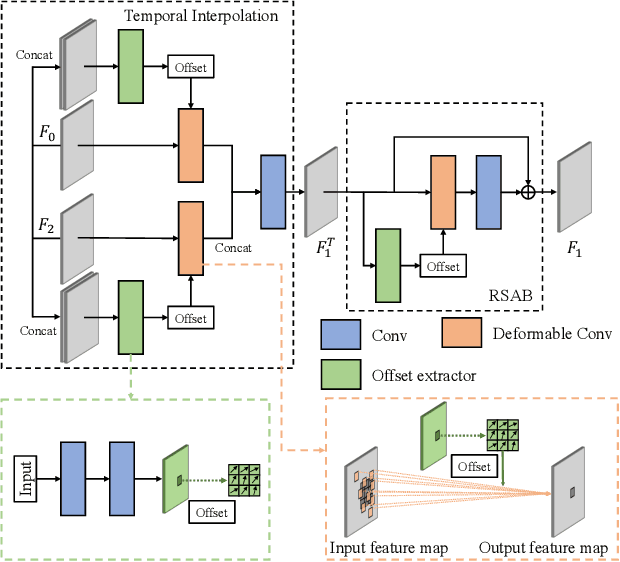
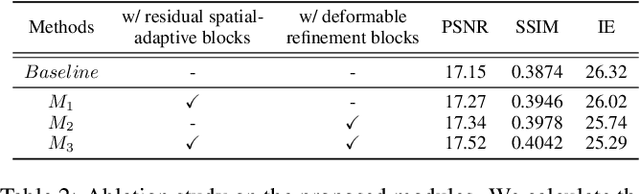
Abstract:Recently, flow-based methods have achieved promising success in video frame interpolation. However, electron microscopic (EM) images suffer from unstable image quality, low PSNR, and disorderly deformation. Existing flow-based interpolation methods cannot precisely compute optical flow for EM images since only predicting each position's unique offset. To overcome these problems, we propose a novel interpolation framework for EM images that progressively synthesizes interpolated features in a coarse-to-fine manner. First, we extract missing intermediate features by the proposed temporal spatial-adaptive (TSA) interpolation module. The TSA interpolation module aggregates temporal contexts and then adaptively samples the spatial-related features with the proposed residual spatial adaptive block. Second, we introduce a stacked deformable refinement block (SDRB) further enhance the reconstruction quality, which is aware of the matching positions and relevant features from input frames with the feedback mechanism. Experimental results demonstrate the superior performance of our approach compared to previous works, both quantitatively and qualitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge