Kyeongtak Han
Solvent: A Framework for Protein Folding
Jul 31, 2023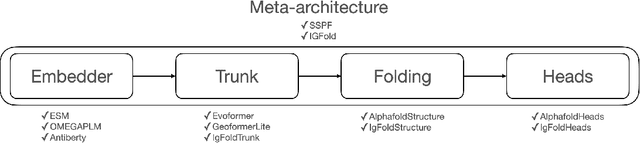

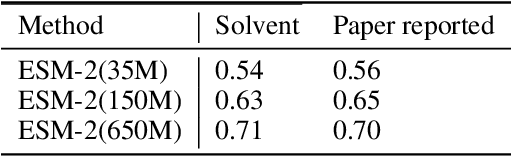
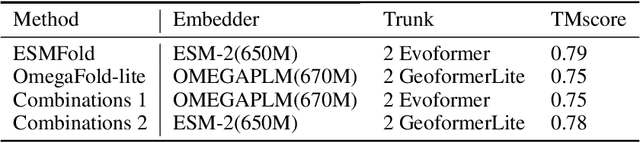
Abstract:Consistency and reliability are crucial for conducting AI research. Many famous research fields, such as object detection, have been compared and validated with solid benchmark frameworks. After AlphaFold2, the protein folding task has entered a new phase, and many methods are proposed based on the component of AlphaFold2. The importance of a unified research framework in protein folding contains implementations and benchmarks to consistently and fairly compare various approaches. To achieve this, we present Solvent, a protein folding framework that supports significant components of state-of-the-art models in the manner of an off-the-shelf interface Solvent contains different models implemented in a unified codebase and supports training and evaluation for defined models on the same dataset. We benchmark well-known algorithms and their components and provide experiments that give helpful insights into the protein structure modeling field. We hope that Solvent will increase the reliability and consistency of proposed models and give efficiency in both speed and costs, resulting in acceleration on protein folding modeling research. The code is available at https://github.com/kakaobrain/solvent, and the project will continue to be developed.
Loss-based Sequential Learning for Active Domain Adaptation
Apr 25, 2022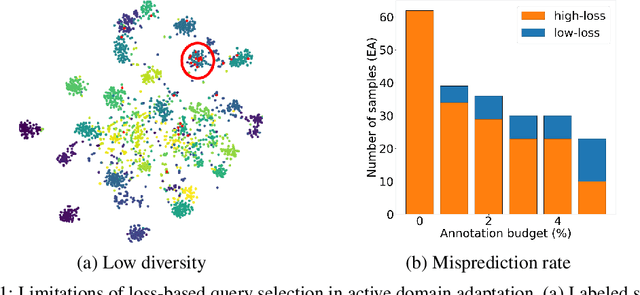



Abstract:Active domain adaptation (ADA) studies have mainly addressed query selection while following existing domain adaptation strategies. However, we argue that it is critical to consider not only query selection criteria but also domain adaptation strategies designed for ADA scenarios. This paper introduces sequential learning considering both domain type (source/target) or labelness (labeled/unlabeled). We first train our model only on labeled target samples obtained by loss-based query selection. When loss-based query selection is applied under domain shift, unuseful high-loss samples gradually increase, and the labeled-sample diversity becomes low. To solve these, we fully utilize pseudo labels of the unlabeled target domain by leveraging loss prediction. We further encourage pseudo labels to have low self-entropy and diverse class distributions. Our model significantly outperforms previous methods as well as baseline models in various benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge