Krithik Ramesh
Broad Institute of MIT and Harvard, Massachusetts Institute of Technology
Lyra: An Efficient and Expressive Subquadratic Architecture for Modeling Biological Sequences
Mar 20, 2025Abstract:Deep learning architectures such as convolutional neural networks and Transformers have revolutionized biological sequence modeling, with recent advances driven by scaling up foundation and task-specific models. The computational resources and large datasets required, however, limit their applicability in biological contexts. We introduce Lyra, a subquadratic architecture for sequence modeling, grounded in the biological framework of epistasis for understanding sequence-to-function relationships. Mathematically, we demonstrate that state space models efficiently capture global epistatic interactions and combine them with projected gated convolutions for modeling local relationships. We demonstrate that Lyra is performant across over 100 wide-ranging biological tasks, achieving state-of-the-art (SOTA) performance in many key areas, including protein fitness landscape prediction, biophysical property prediction (e.g. disordered protein region functions) peptide engineering applications (e.g. antibody binding, cell-penetrating peptide prediction), RNA structure analysis, RNA function prediction, and CRISPR guide design. It achieves this with orders-of-magnitude improvements in inference speed and reduction in parameters (up to 120,000-fold in our tests) compared to recent biology foundation models. Using Lyra, we were able to train and run every task in this study on two or fewer GPUs in under two hours, democratizing access to biological sequence modeling at SOTA performance, with potential applications to many fields.
LoLCATs: On Low-Rank Linearizing of Large Language Models
Oct 14, 2024
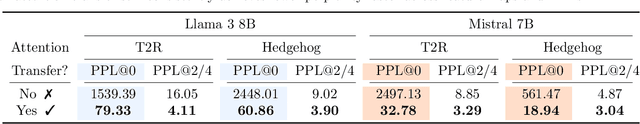
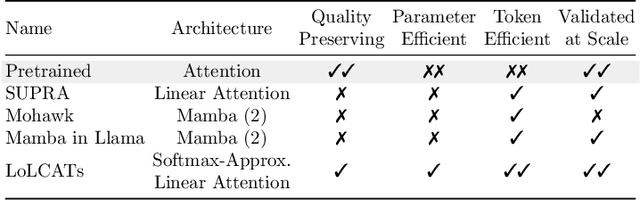
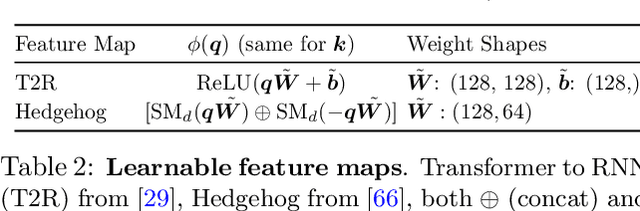
Abstract:Recent works show we can linearize large language models (LLMs) -- swapping the quadratic attentions of popular Transformer-based LLMs with subquadratic analogs, such as linear attention -- avoiding the expensive pretraining costs. However, linearizing LLMs often significantly degrades model quality, still requires training over billions of tokens, and remains limited to smaller 1.3B to 7B LLMs. We thus propose Low-rank Linear Conversion via Attention Transfer (LoLCATs), a simple two-step method that improves LLM linearizing quality with orders of magnitudes less memory and compute. We base these steps on two findings. First, we can replace an LLM's softmax attentions with closely-approximating linear attentions, simply by training the linear attentions to match their softmax counterparts with an output MSE loss ("attention transfer"). Then, this enables adjusting for approximation errors and recovering LLM quality simply with low-rank adaptation (LoRA). LoLCATs significantly improves linearizing quality, training efficiency, and scalability. We significantly reduce the linearizing quality gap and produce state-of-the-art subquadratic LLMs from Llama 3 8B and Mistral 7B v0.1, leading to 20+ points of improvement on 5-shot MMLU. Furthermore, LoLCATs does so with only 0.2% of past methods' model parameters and 0.4% of their training tokens. Finally, we apply LoLCATs to create the first linearized 70B and 405B LLMs (50x larger than prior work). When compared with prior approaches under the same compute budgets, LoLCATs significantly improves linearizing quality, closing the gap between linearized and original Llama 3.1 70B and 405B LLMs by 77.8% and 78.1% on 5-shot MMLU.
PCN: A Deep Learning Approach to Jet Tagging Utilizing Novel Graph Construction Methods and Chebyshev Graph Convolutions
Sep 12, 2023Abstract:Jet tagging is a classification problem in high-energy physics experiments that aims to identify the collimated sprays of subatomic particles, jets, from particle collisions and tag them to their emitter particle. Advances in jet tagging present opportunities for searches of new physics beyond the Standard Model. Current approaches use deep learning to uncover hidden patterns in complex collision data. However, the representation of jets as inputs to a deep learning model have been varied, and often, informative features are withheld from models. In this study, we propose a graph-based representation of a jet that encodes the most information possible. To learn best from this representation, we design Particle Chebyshev Network (PCN), a graph neural network (GNN) using Chebyshev graph convolutions (ChebConv). ChebConv has been demonstrated as an effective alternative to classical graph convolutions in GNNs and has yet to be explored in jet tagging. PCN achieves a substantial improvement in accuracy over existing taggers and opens the door to future studies into graph-based representations of jets and ChebConv layers in high-energy physics experiments. Code is available at https://github.com/YVSemlani/PCN-Jet-Tagging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge