Kim L. Boyer
Semantic Context Forests for Learning-Based Knee Cartilage Segmentation in 3D MR Images
Apr 22, 2014
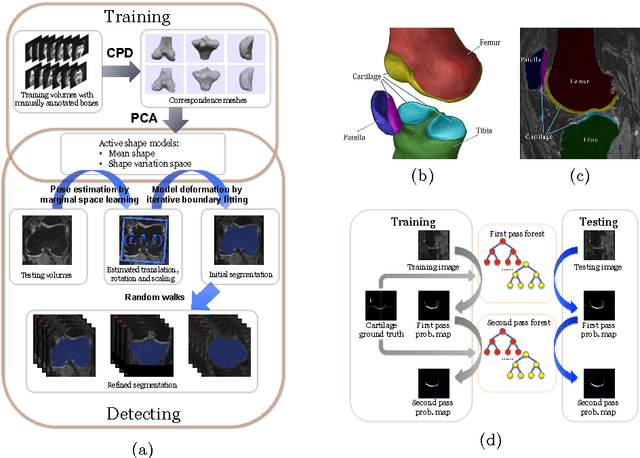
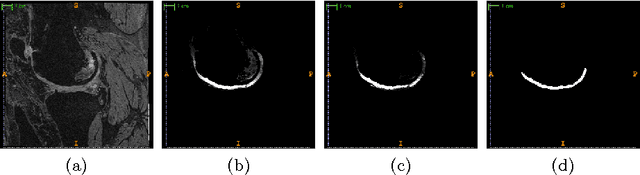
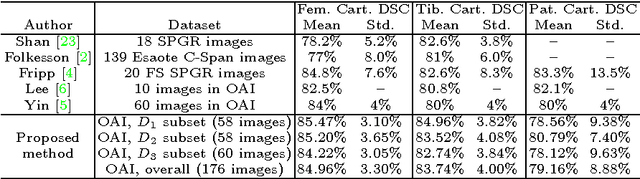
Abstract:The automatic segmentation of human knee cartilage from 3D MR images is a useful yet challenging task due to the thin sheet structure of the cartilage with diffuse boundaries and inhomogeneous intensities. In this paper, we present an iterative multi-class learning method to segment the femoral, tibial and patellar cartilage simultaneously, which effectively exploits the spatial contextual constraints between bone and cartilage, and also between different cartilages. First, based on the fact that the cartilage grows in only certain area of the corresponding bone surface, we extract the distance features of not only to the surface of the bone, but more informatively, to the densely registered anatomical landmarks on the bone surface. Second, we introduce a set of iterative discriminative classifiers that at each iteration, probability comparison features are constructed from the class confidence maps derived by previously learned classifiers. These features automatically embed the semantic context information between different cartilages of interest. Validated on a total of 176 volumes from the Osteoarthritis Initiative (OAI) dataset, the proposed approach demonstrates high robustness and accuracy of segmentation in comparison with existing state-of-the-art MR cartilage segmentation methods.
Feature Learning by Multidimensional Scaling and its Applications in Object Recognition
Jun 14, 2013
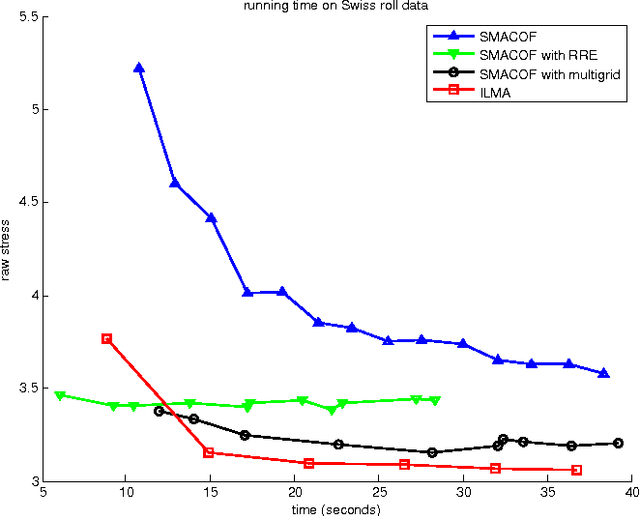


Abstract:We present the MDS feature learning framework, in which multidimensional scaling (MDS) is applied on high-level pairwise image distances to learn fixed-length vector representations of images. The aspects of the images that are captured by the learned features, which we call MDS features, completely depend on what kind of image distance measurement is employed. With properly selected semantics-sensitive image distances, the MDS features provide rich semantic information about the images that is not captured by other feature extraction techniques. In our work, we introduce the iterated Levenberg-Marquardt algorithm for solving MDS, and study the MDS feature learning with IMage Euclidean Distance (IMED) and Spatial Pyramid Matching (SPM) distance. We present experiments on both synthetic data and real images --- the publicly accessible UIUC car image dataset. The MDS features based on SPM distance achieve exceptional performance for the car recognition task.
Tracking Tetrahymena Pyriformis Cells using Decision Trees
Jul 13, 2012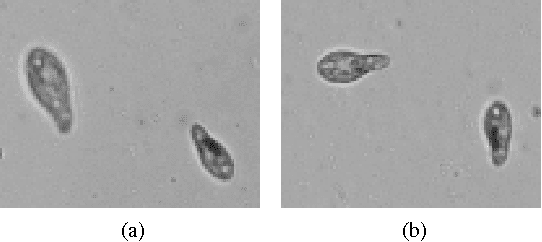
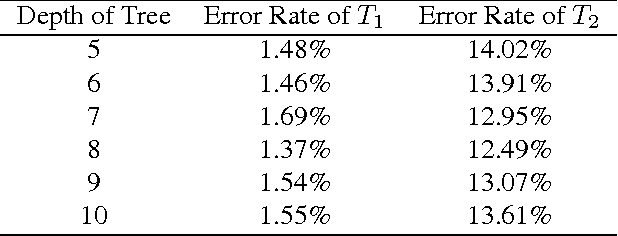
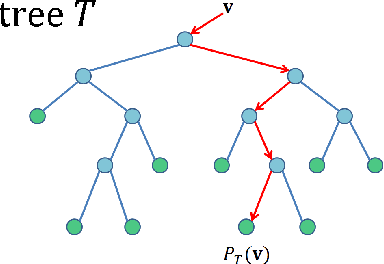
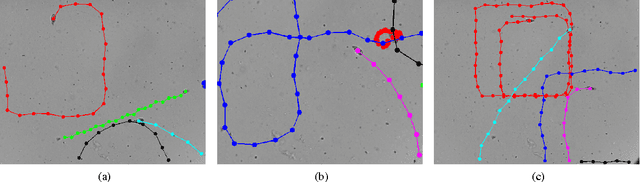
Abstract:Matching cells over time has long been the most difficult step in cell tracking. In this paper, we approach this problem by recasting it as a classification problem. We construct a feature set for each cell, and compute a feature difference vector between a cell in the current frame and a cell in a previous frame. Then we determine whether the two cells represent the same cell over time by training decision trees as our binary classifiers. With the output of decision trees, we are able to formulate an assignment problem for our cell association task and solve it using a modified version of the Hungarian algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge