Katharina Nöh
EAP4EMSIG -- Enhancing Event-Driven Microscopy for Microfluidic Single-Cell Analysis
Mar 30, 2025Abstract:Microfluidic Live-Cell Imaging yields data on microbial cell factories. However, continuous acquisition is challenging as high-throughput experiments often lack realtime insights, delaying responses to stochastic events. We introduce three components in the Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cell Analysis: a fast, accurate Deep Learning autofocusing method predicting the focus offset, an evaluation of real-time segmentation methods and a realtime data analysis dashboard. Our autofocusing achieves a Mean Absolute Error of 0.0226\textmu m with inference times below 50~ms. Among eleven Deep Learning segmentation methods, Cellpose~3 reached a Panoptic Quality of 93.58\%, while a distance-based method is fastest (121~ms, Panoptic Quality 93.02\%). All six Deep Learning Foundation Models were unsuitable for real-time segmentation.
PyUAT: Open-source Python framework for efficient and scalable cell tracking
Mar 27, 2025Abstract:Tracking individual cells in live-cell imaging provides fundamental insights, inevitable for studying causes and consequences of phenotypic heterogeneity, responses to changing environmental conditions or stressors. Microbial cell tracking, characterized by stochastic cell movements and frequent cell divisions, remains a challenging task when imaging frame rates must be limited to avoid counterfactual results. A promising way to overcome this limitation is uncertainty-aware tracking (UAT), which uses statistical models, calibrated to empirically observed cell behavior, to predict likely cell associations. We present PyUAT, an efficient and modular Python implementation of UAT for tracking microbial cells in time-lapse imaging. We demonstrate its performance on a large 2D+t data set and investigate the influence of modular biological models and imaging intervals on the tracking performance. The open-source PyUAT software is available at https://github.com/JuBiotech/PyUAT, including example notebooks for immediate use in Google Colab.
How To Make Your Cell Tracker Say "I dunno!"
Mar 12, 2025Abstract:Cell tracking is a key computational task in live-cell microscopy, but fully automated analysis of high-throughput imaging requires reliable and, thus, uncertainty-aware data analysis tools, as the amount of data recorded within a single experiment exceeds what humans are able to overlook. We here propose and benchmark various methods to reason about and quantify uncertainty in linear assignment-based cell tracking algorithms. Our methods take inspiration from statistics and machine learning, leveraging two perspectives on the cell tracking problem explored throughout this work: Considering it as a Bayesian inference problem and as a classification problem. Our methods admit a framework-like character in that they equip any frame-to-frame tracking method with uncertainty quantification. We demonstrate this by applying it to various existing tracking algorithms including the recently presented Transformer-based trackers. We demonstrate empirically that our methods yield useful and well-calibrated tracking uncertainties.
EAP4EMSIG -- Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cells Analysis
Nov 06, 2024Abstract:Microfluidic Live-Cell Imaging (MLCI) generates high-quality data that allows biotechnologists to study cellular growth dynamics in detail. However, obtaining these continuous data over extended periods is challenging, particularly in achieving accurate and consistent real-time event classification at the intersection of imaging and stochastic biology. To address this issue, we introduce the Experiment Automation Pipeline for Event-Driven Microscopy to Smart Microfluidic Single-Cells Analysis (EAP4EMSIG). In particular, we present initial zero-shot results from the real-time segmentation module of our approach. Our findings indicate that among four State-Of-The- Art (SOTA) segmentation methods evaluated, Omnipose delivers the highest Panoptic Quality (PQ) score of 0.9336, while Contour Proposal Network (CPN) achieves the fastest inference time of 185 ms with the second-highest PQ score of 0.8575. Furthermore, we observed that the vision foundation model Segment Anything is unsuitable for this particular use case.
Parameter-efficient Bayesian Neural Networks for Uncertainty-aware Depth Estimation
Sep 25, 2024



Abstract:State-of-the-art computer vision tasks, like monocular depth estimation (MDE), rely heavily on large, modern Transformer-based architectures. However, their application in safety-critical domains demands reliable predictive performance and uncertainty quantification. While Bayesian neural networks provide a conceptually simple approach to serve those requirements, they suffer from the high dimensionality of the parameter space. Parameter-efficient fine-tuning (PEFT) methods, in particular low-rank adaptations (LoRA), have emerged as a popular strategy for adapting large-scale models to down-stream tasks by performing parameter inference on lower-dimensional subspaces. In this work, we investigate the suitability of PEFT methods for subspace Bayesian inference in large-scale Transformer-based vision models. We show that, indeed, combining BitFit, DiffFit, LoRA, and CoLoRA, a novel LoRA-inspired PEFT method, with Bayesian inference enables more robust and reliable predictive performance in MDE.
Cell tracking for live-cell microscopy using an activity-prioritized assignment strategy
Oct 20, 2022
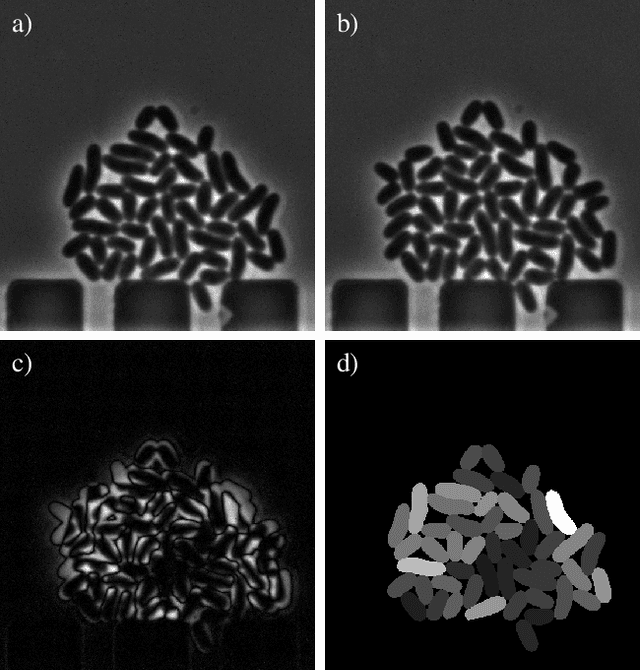
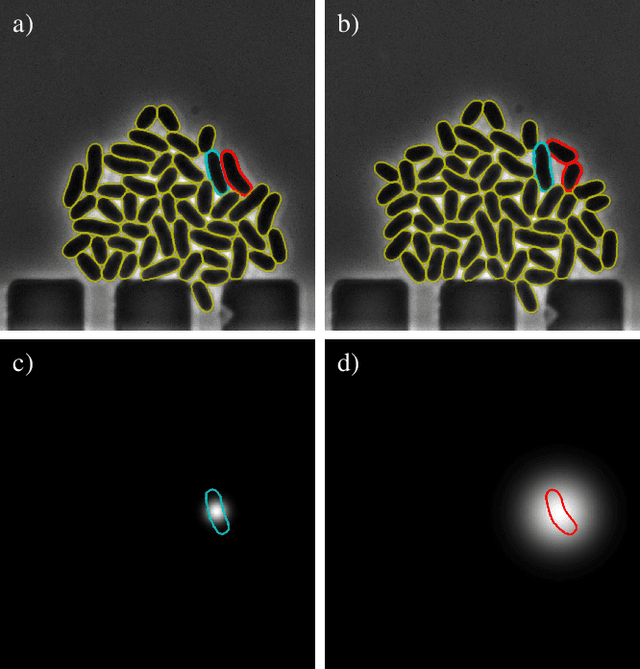
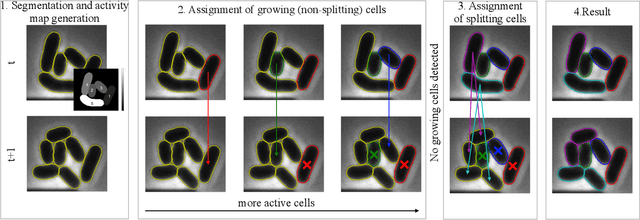
Abstract:Cell tracking is an essential tool in live-cell imaging to determine single-cell features, such as division patterns or elongation rates. Unlike in common multiple object tracking, in microbial live-cell experiments cells are growing, moving, and dividing over time, to form cell colonies that are densely packed in mono-layer structures. With increasing cell numbers, following the precise cell-cell associations correctly over many generations becomes more and more challenging, due to the massively increasing number of possible associations. To tackle this challenge, we propose a fast parameter-free cell tracking approach, which consists of activity-prioritized nearest neighbor assignment of growing cells and a combinatorial solver that assigns splitting mother cells to their daughters. As input for the tracking, Omnipose is utilized for instance segmentation. Unlike conventional nearest-neighbor-based tracking approaches, the assignment steps of our proposed method are based on a Gaussian activity-based metric, predicting the cell-specific migration probability, thereby limiting the number of erroneous assignments. In addition to being a building block for cell tracking, the proposed activity map is a standalone tracking-free metric for indicating cell activity. Finally, we perform a quantitative analysis of the tracking accuracy for different frame rates, to inform life scientists about a suitable (in terms of tracking performance) choice of the frame rate for their cultivation experiments, when cell tracks are the desired key outcome.
Automated Characterization of Catalytically Active Inclusion Body Production in Biotechnological Screening Systems
Sep 30, 2022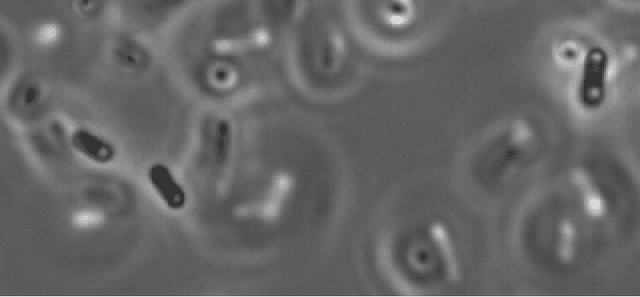
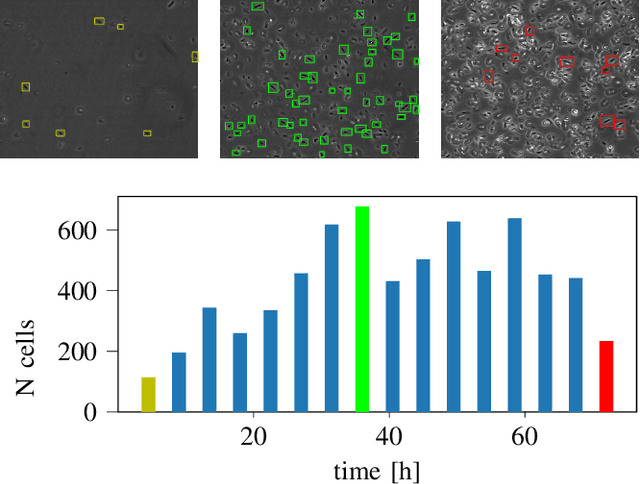
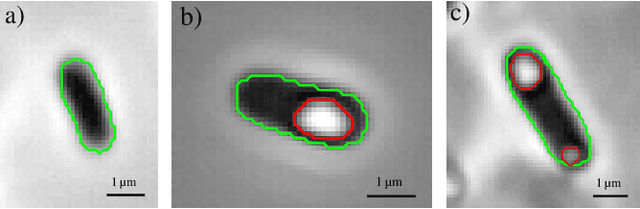
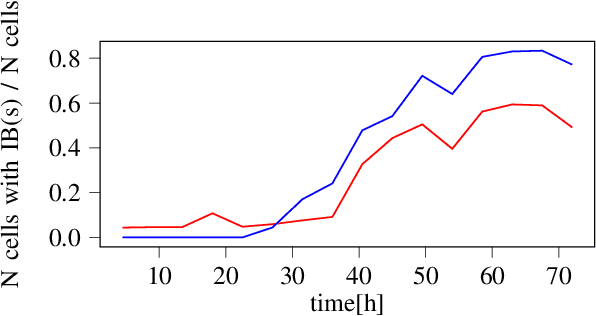
Abstract:We here propose an automated pipeline for the microscopy image-based characterization of catalytically active inclusion bodies (CatIBs), which includes a fully automatic experimental high-throughput workflow combined with a hybrid approach for multi-object microbial cell segmentation. For automated microscopy, a CatIB producer strain was cultivated in a microbioreactor from which samples were injected into a flow chamber. The flow chamber was fixed under a microscope and an integrated camera took a series of images per sample. To explore heterogeneity of CatIB development during the cultivation and track the size and quantity of CatIBs over time, a hybrid image processing pipeline approach was developed, which combines an ML-based detection of in-focus cells with model-based segmentation. The experimental setup in combination with an automated image analysis unlocks high-throughput screening of CatIB production, saving time and resources. Biotechnological relevance - CatIBs have wide application in synthetic chemistry and biocatalysis, but also could have future biomedical applications such as therapeutics. The proposed hybrid automatic image processing pipeline can be adjusted to treat comparable biological microorganisms, where fully data-driven ML-based segmentation approaches are not feasible due to the lack of training data. Our work is the first step towards image-based bioprocess control.
A hybrid multi-object segmentation framework with model-based B-splines for microbial single cell analysis
May 03, 2022
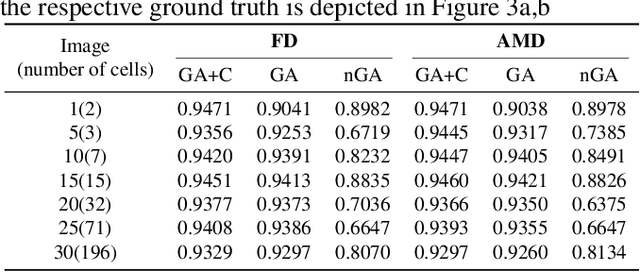

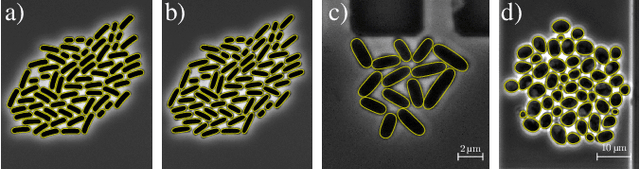
Abstract:In this paper, we propose a hybrid approach for multi-object microbial cell segmentation. The approach combines an ML-based detection with a geometry-aware variational-based segmentation using B-splines that are parametrized based on a geometric model of the cell shape. The detection is done first using YOLOv5. In a second step, each detected cell is segmented individually. Thus, the segmentation only needs to be done on a per-cell basis, which makes it amenable to a variational approach that incorporates prior knowledge on the geometry. Here, the contour of the segmentation is modelled as closed uniform cubic B-spline, whose control points are parametrized using the known cell geometry. Compared to purely ML-based segmentation approaches, which need accurate segmentation maps as training data that are very laborious to produce, our method just needs bounding boxes as training data. Still, the proposed method performs on par with ML-based segmentation approaches usually used in this context. We study the performance of the proposed method on time-lapse microscopy data of Corynebacterium glutamicum.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge