Benjamin Berkels
A Generalization Bound for a Family of Implicit Networks
Oct 09, 2024

Abstract:Implicit networks are a class of neural networks whose outputs are defined by the fixed point of a parameterized operator. They have enjoyed success in many applications including natural language processing, image processing, and numerous other applications. While they have found abundant empirical success, theoretical work on its generalization is still under-explored. In this work, we consider a large family of implicit networks defined parameterized contractive fixed point operators. We show a generalization bound for this class based on a covering number argument for the Rademacher complexity of these architectures.
Direct Motif Extraction from High Resolution Crystalline STEM Images
Mar 13, 2023Abstract:During the last decade, automatic data analysis methods concerning different aspects of crystal analysis have been developed, e.g., unsupervised primitive unit cell extraction and automated crystal distortion and defects detection. However, an automatic, unsupervised motif extraction method is still not widely available yet. Here, we propose and demonstrate a novel method for the automatic motif extraction in real space from crystalline images based on a variational approach involving the unit cell projection operator. Due to the non-convex nature of the resulting minimization problem, a multi-stage algorithm is used. First, we determine the primitive unit cell in form of two lattice vectors. Second, a motif image is estimated using the unit cell information. Finally, the motif is determined in terms of atom positions inside the unit cell. The method was tested on various synthetic and experimental HAADF STEM images. The results are a representation of the motif in form of an image, atomic positions, primitive unit cell vectors, and a denoised and a modeled reconstruction of the input image. The method was applied to extract the primitive cells of complex $\mu$-phase structures Nb$_\text{6.4}$Co$_\text{6.6}$ and Nb$_\text{7}$Co$_\text{6}$, where subtle differences between their interplanar spacings were determined.
Cell tracking for live-cell microscopy using an activity-prioritized assignment strategy
Oct 20, 2022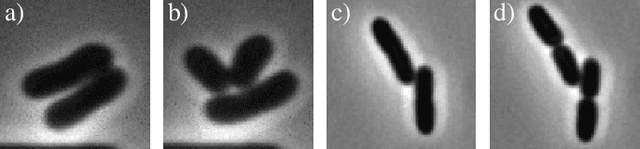
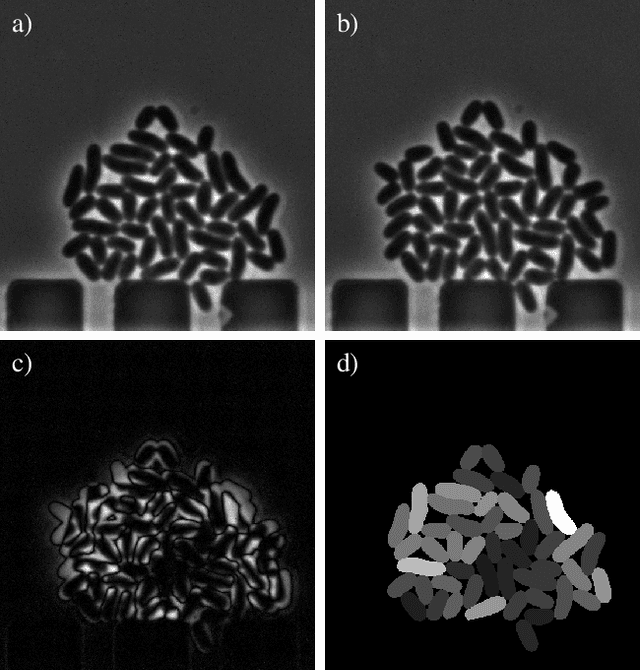
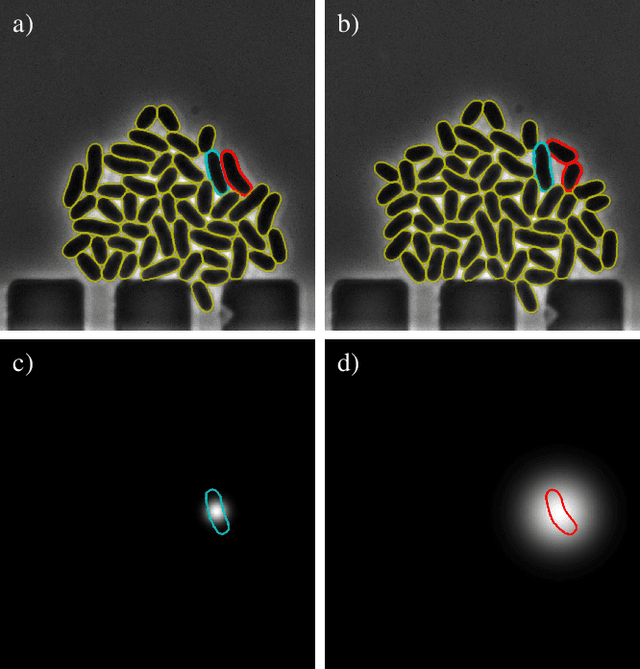
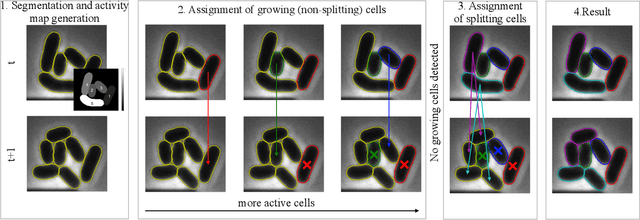
Abstract:Cell tracking is an essential tool in live-cell imaging to determine single-cell features, such as division patterns or elongation rates. Unlike in common multiple object tracking, in microbial live-cell experiments cells are growing, moving, and dividing over time, to form cell colonies that are densely packed in mono-layer structures. With increasing cell numbers, following the precise cell-cell associations correctly over many generations becomes more and more challenging, due to the massively increasing number of possible associations. To tackle this challenge, we propose a fast parameter-free cell tracking approach, which consists of activity-prioritized nearest neighbor assignment of growing cells and a combinatorial solver that assigns splitting mother cells to their daughters. As input for the tracking, Omnipose is utilized for instance segmentation. Unlike conventional nearest-neighbor-based tracking approaches, the assignment steps of our proposed method are based on a Gaussian activity-based metric, predicting the cell-specific migration probability, thereby limiting the number of erroneous assignments. In addition to being a building block for cell tracking, the proposed activity map is a standalone tracking-free metric for indicating cell activity. Finally, we perform a quantitative analysis of the tracking accuracy for different frame rates, to inform life scientists about a suitable (in terms of tracking performance) choice of the frame rate for their cultivation experiments, when cell tracks are the desired key outcome.
Automated Characterization of Catalytically Active Inclusion Body Production in Biotechnological Screening Systems
Sep 30, 2022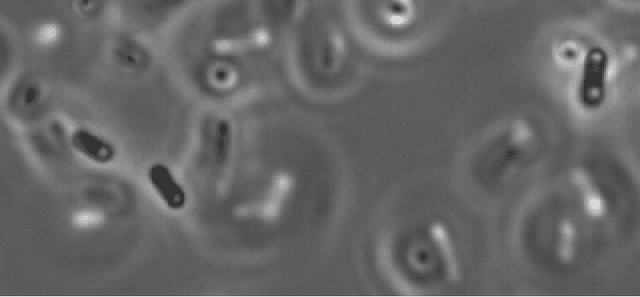
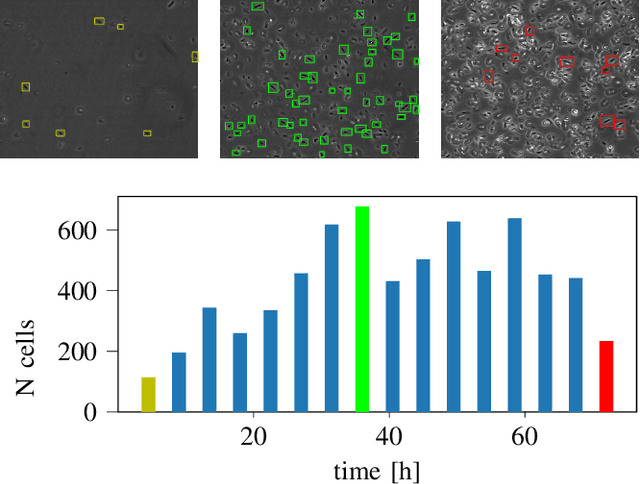
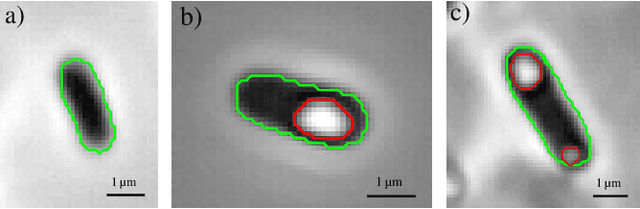
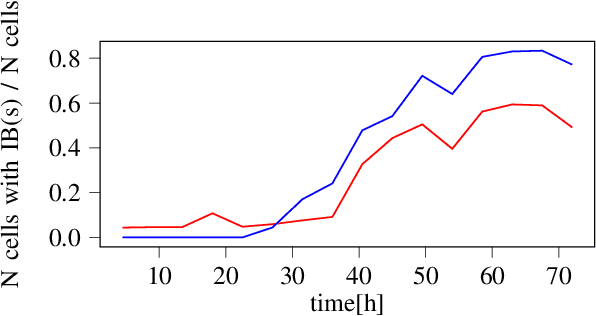
Abstract:We here propose an automated pipeline for the microscopy image-based characterization of catalytically active inclusion bodies (CatIBs), which includes a fully automatic experimental high-throughput workflow combined with a hybrid approach for multi-object microbial cell segmentation. For automated microscopy, a CatIB producer strain was cultivated in a microbioreactor from which samples were injected into a flow chamber. The flow chamber was fixed under a microscope and an integrated camera took a series of images per sample. To explore heterogeneity of CatIB development during the cultivation and track the size and quantity of CatIBs over time, a hybrid image processing pipeline approach was developed, which combines an ML-based detection of in-focus cells with model-based segmentation. The experimental setup in combination with an automated image analysis unlocks high-throughput screening of CatIB production, saving time and resources. Biotechnological relevance - CatIBs have wide application in synthetic chemistry and biocatalysis, but also could have future biomedical applications such as therapeutics. The proposed hybrid automatic image processing pipeline can be adjusted to treat comparable biological microorganisms, where fully data-driven ML-based segmentation approaches are not feasible due to the lack of training data. Our work is the first step towards image-based bioprocess control.
A hybrid multi-object segmentation framework with model-based B-splines for microbial single cell analysis
May 03, 2022
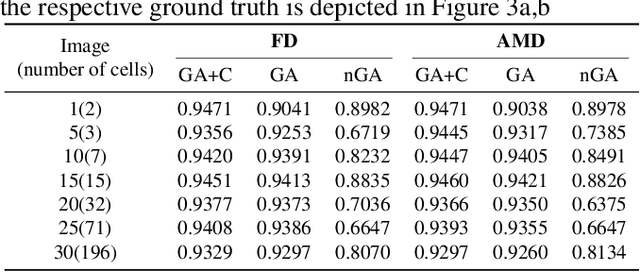

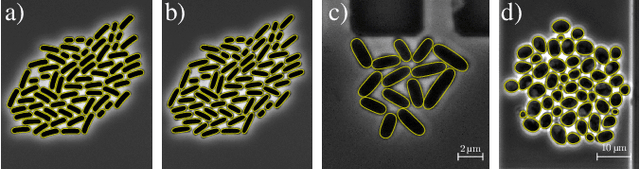
Abstract:In this paper, we propose a hybrid approach for multi-object microbial cell segmentation. The approach combines an ML-based detection with a geometry-aware variational-based segmentation using B-splines that are parametrized based on a geometric model of the cell shape. The detection is done first using YOLOv5. In a second step, each detected cell is segmented individually. Thus, the segmentation only needs to be done on a per-cell basis, which makes it amenable to a variational approach that incorporates prior knowledge on the geometry. Here, the contour of the segmentation is modelled as closed uniform cubic B-spline, whose control points are parametrized using the known cell geometry. Compared to purely ML-based segmentation approaches, which need accurate segmentation maps as training data that are very laborious to produce, our method just needs bounding boxes as training data. Still, the proposed method performs on par with ML-based segmentation approaches usually used in this context. We study the performance of the proposed method on time-lapse microscopy data of Corynebacterium glutamicum.
A distribution-dependent Mumford-Shah model for unsupervised hyperspectral image segmentation
Mar 28, 2022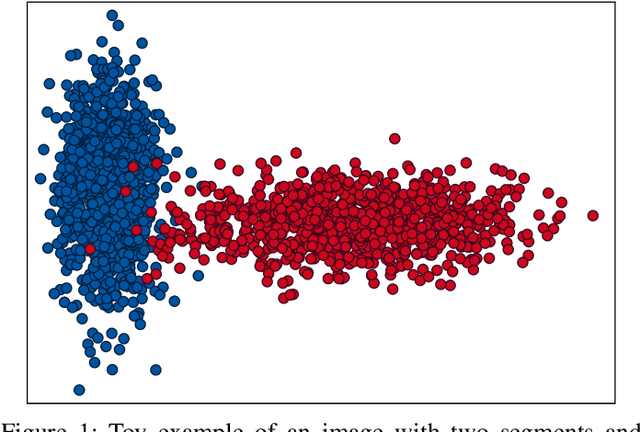
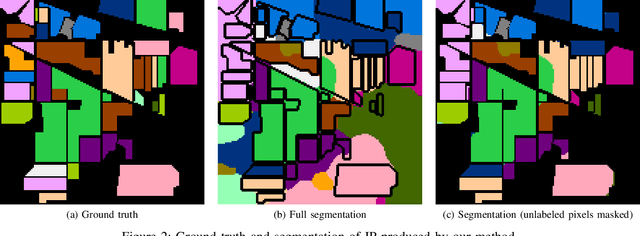
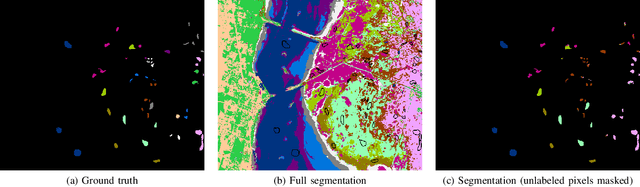
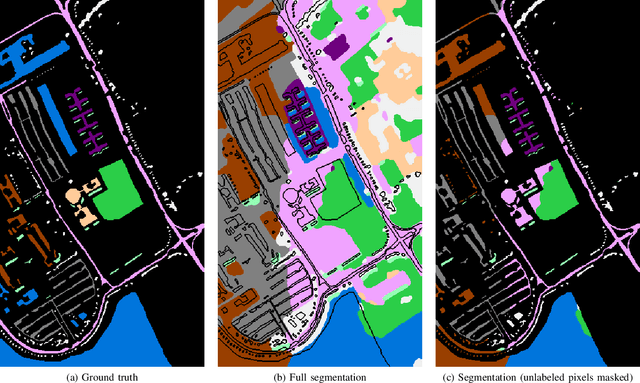
Abstract:Hyperspectral images provide a rich representation of the underlying spectrum for each pixel, allowing for a pixel-wise classification/segmentation into different classes. As the acquisition of labeled training data is very time-consuming, unsupervised methods become crucial in hyperspectral image analysis. The spectral variability and noise in hyperspectral data make this task very challenging and define special requirements for such methods. Here, we present a novel unsupervised hyperspectral segmentation framework. It starts with a denoising and dimensionality reduction step by the well-established Minimum Noise Fraction (MNF) transform. Then, the Mumford-Shah (MS) segmentation functional is applied to segment the data. We equipped the MS functional with a novel robust distribution-dependent indicator function designed to handle the characteristic challenges of hyperspectral data. To optimize our objective function with respect to the parameters for which no closed form solution is available, we propose an efficient fixed point iteration scheme. Numerical experiments on four public benchmark datasets show that our method produces competitive results, which outperform two state-of-the-art methods substantially on three of these datasets.
Automation of Hemocompatibility Analysis Using Image Segmentation and a Random Forest
Oct 13, 2020



Abstract:The hemocompatibility of blood-contacting medical devices remains one of the major challenges in biomedical engineering and makes research in the field of new and improved materials inevitable. However, current in-vitro test and analysis methods are still lacking standardization and comparability, which impedes advances in material design. For example, the optical platelet analysis of material in-vitro hemocompatibility tests is carried out manually or semi-manually by each research group individually. As a step towards standardization, this paper proposes an automation approach for the optical platelet count and analysis. To this end, fluorescence images are segmented using Zach's convexification of the multiphase-phase piecewise constant Mumford--Shah model. The resulting connected components of the non-background segments then need to be classified as platelet or no platelet. Therefore, a supervised random forest is applied to feature vectors derived from the components using features like area, perimeter and circularity. With an overall high accuracy and low error rates, the random forest achieves reliable results. This is supported by high areas under the receiver-operator and the prediction-recall curve, respectively. We developed a new method for a fast, user-independent and reproducible analysis of material hemocompatibility tests, which is therefore a unique and powerful tool for advances in biomaterial research.
Variational Multi-Phase Segmentation using High-Dimensional Local Features
Feb 26, 2019



Abstract:We propose a novel method for multi-phase segmentation of images based on high-dimensional local feature vectors. While the method was developed for the segmentation of extremely noisy crystal images based on localized Fourier transforms, the resulting framework is not tied to specific feature descriptors. For instance, using local spectral histograms as features, it allows for robust texture segmentation. The segmentation itself is based on the multi-phase Mumford-Shah model. Initializing the high-dimensional mean features directly is computationally too demanding and ill-posed in practice. This is resolved by projecting the features onto a low-dimensional space using principle component analysis. The resulting objective functional is minimized using a convexification and the Chambolle-Pock algorithm. Numerical results are presented, illustrating that the algorithm is very competitive in texture segmentation with state-of-the-art performance on the Prague benchmark and provides new possibilities in crystal segmentation, being robust to extreme noise and requiring no prior knowledge of the crystal structure.
Mesh-to-raster based non-rigid registration of multi-modal images
Oct 31, 2017Abstract:Region of interest (ROI) alignment in medical images plays a crucial role in diagnostics, procedure planning, treatment, and follow-up. Frequently, a model is represented as triangulated mesh while the patient data is provided from CAT scanners as pixel or voxel data. Previously, we presented a 2D method for curve-to-pixel registration. This paper contributes (i) a general mesh-to-raster (M2R) framework to register ROIs in multi-modal images; (ii) a 3D surface-to-voxel application, and (iii) a comprehensive quantitative evaluation in 2D using ground truth provided by the simultaneous truth and performance level estimation (STAPLE) method. The registration is formulated as a minimization problem where the objective consists of a data term, which involves the signed distance function of the ROI from the reference image, and a higher order elastic regularizer for the deformation. The evaluation is based on quantitative light-induced fluoroscopy (QLF) and digital photography (DP) of decalcified teeth. STAPLE is computed on 150 image pairs from 32 subjects, each showing one corresponding tooth in both modalities. The ROI in each image is manually marked by three experts (900 curves in total). In the QLF-DP setting, our approach significantly outperforms the mutual information-based registration algorithm implemented with the Insight Segmentation and Registration Toolkit (ITK) and Elastix.
Joint denoising and distortion correction of atomic scale scanning transmission electron microscopy images
Dec 24, 2016
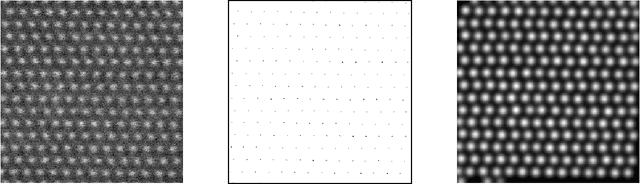


Abstract:Nowadays, modern electron microscopes deliver images at atomic scale. The precise atomic structure encodes information about material properties. Thus, an important ingredient in the image analysis is to locate the centers of the atoms shown in micrographs as precisely as possible. Here, we consider scanning transmission electron microscopy (STEM), which acquires data in a rastering pattern, pixel by pixel. Due to this rastering combined with the magnification to atomic scale, movements of the specimen even at the nanometer scale lead to random image distortions that make precise atom localization difficult. Given a series of STEM images, we derive a Bayesian method that jointly estimates the distortion in each image and reconstructs the underlying atomic grid of the material by fitting the atom bumps with suitable bump functions. The resulting highly non-convex minimization problems are solved numerically with a trust region approach. Well-posedness of the reconstruction method and the model behavior for faster and faster rastering are investigated using variational techniques. The performance of the method is finally evaluated on both synthetic and real experimental data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge