Jules Dedieu
Tribvn Healthcare
Monte Carlo Augmented Actor-Critic for Sparse Reward Deep Reinforcement Learning from Suboptimal Demonstrations
Oct 14, 2022
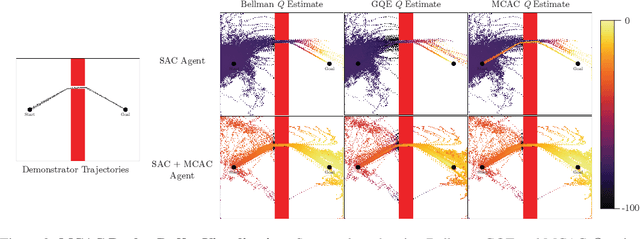

Abstract:Providing densely shaped reward functions for RL algorithms is often exceedingly challenging, motivating the development of RL algorithms that can learn from easier-to-specify sparse reward functions. This sparsity poses new exploration challenges. One common way to address this problem is using demonstrations to provide initial signal about regions of the state space with high rewards. However, prior RL from demonstrations algorithms introduce significant complexity and many hyperparameters, making them hard to implement and tune. We introduce Monte Carlo Augmented Actor Critic (MCAC), a parameter free modification to standard actor-critic algorithms which initializes the replay buffer with demonstrations and computes a modified $Q$-value by taking the maximum of the standard temporal distance (TD) target and a Monte Carlo estimate of the reward-to-go. This encourages exploration in the neighborhood of high-performing trajectories by encouraging high $Q$-values in corresponding regions of the state space. Experiments across $5$ continuous control domains suggest that MCAC can be used to significantly increase learning efficiency across $6$ commonly used RL and RL-from-demonstrations algorithms. See https://sites.google.com/view/mcac-rl for code and supplementary material.
Robust Mitosis Detection Using a Cascade Mask-RCNN Approach With Domain-Specific Residual Cycle-GAN Data Augmentation
Sep 28, 2021
Abstract:For the MIDOG mitosis detection challenge, we created a cascade algorithm consisting of a Mask-RCNN detector, followed by a classification ensemble consisting of ResNet50 and DenseNet201 to refine detected mitotic candidates. The MIDOG training data consists of 200 frames originating from four scanners, three of which are annotated for mitotic instances with centroid annotations. Our main algorithmic choices are as follows: first, to enhance the generalizability of our detector and classification networks, we use a state-of-the-art residual Cycle-GAN to transform each scanner domain to every other scanner domain. During training, we then randomly load, for each image, one of the four domains. In this way, our networks can learn from the fourth non-annotated scanner domain even if we don't have annotations for it. Second, for training the detector network, rather than using centroid-based fixed-size bounding boxes, we create mitosis-specific bounding boxes. We do this by manually annotating a small selection of mitoses, training a Mask-RCNN on this small dataset, and applying it to the rest of the data to obtain full annotations. We trained the follow-up classification ensemble using only the challenge-provided positive and hard-negative examples. On the preliminary test set, the algorithm scores an F1 score of 0.7578, putting us as the second-place team on the leaderboard.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge